
Drosophila virilis (Fruit fly)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Eremoneura; Cyclorrhapha; Schizophora; Acalyptratae
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
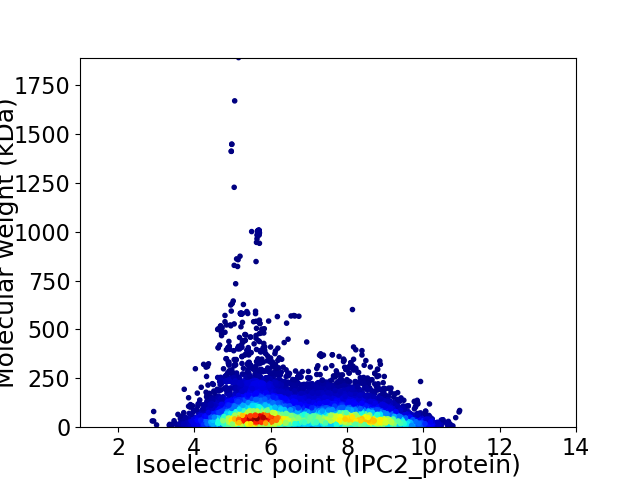
Virtual 2D-PAGE plot for 18316 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B4LSM7|B4LSM7_DROVI Uncharacterized protein isoform A OS=Drosophila virilis OX=7244 GN=Dvir\GJ16490 PE=4 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84LISLVIFCLCVCAAIWPANALQCYY26 pKa = 8.15QCIGSEE32 pKa = 4.5CDD34 pKa = 4.64DD35 pKa = 4.82LDD37 pKa = 3.94DD38 pKa = 5.46SKK40 pKa = 11.8VVSCNLDD47 pKa = 3.39EE48 pKa = 4.46TTSTLTTEE56 pKa = 4.24DD57 pKa = 3.63TTTEE61 pKa = 4.18SLPDD65 pKa = 3.49TSTTSSGSSTGGSTTSPGPTVDD87 pKa = 3.59TPATEE92 pKa = 4.21SSPPSTTDD100 pKa = 3.08QSDD103 pKa = 3.63TTTEE107 pKa = 4.08ATSEE111 pKa = 4.19TTTTGEE117 pKa = 4.02PSEE120 pKa = 4.33STEE123 pKa = 4.12TTEE126 pKa = 4.24TTTGEE131 pKa = 4.13PSGSTEE137 pKa = 4.2TTDD140 pKa = 3.14STTEE144 pKa = 3.95EE145 pKa = 4.29PSGSTEE151 pKa = 4.06TTEE154 pKa = 4.12STTEE158 pKa = 3.87STTDD162 pKa = 3.19STSDD166 pKa = 3.41STSDD170 pKa = 3.44STSDD174 pKa = 3.49STSDD178 pKa = 3.37SSPGSTASSTSEE190 pKa = 3.94STSEE194 pKa = 4.09STSDD198 pKa = 3.43STSDD202 pKa = 3.48STSDD206 pKa = 3.27SSSGSTAGSTSDD218 pKa = 3.41STSEE222 pKa = 4.09STSDD226 pKa = 3.42STSDD230 pKa = 3.19STTGSTSDD238 pKa = 3.48STSDD242 pKa = 3.44STSDD246 pKa = 3.38STSSPEE252 pKa = 3.7SSIPSSSGSTEE263 pKa = 3.86SSTAASTGSSEE274 pKa = 4.4TGEE277 pKa = 4.39STTSGSTEE285 pKa = 3.98SSTSSSASVVNPGNLEE301 pKa = 4.26NIQLALQQAALPGGIRR317 pKa = 11.84SGRR320 pKa = 11.84ARR322 pKa = 11.84RR323 pKa = 11.84SLAAQAADD331 pKa = 3.3QLIRR335 pKa = 11.84NYY337 pKa = 10.52RR338 pKa = 11.84SYY340 pKa = 11.16EE341 pKa = 3.86FNGRR345 pKa = 11.84ATNYY349 pKa = 7.59RR350 pKa = 11.84TTACYY355 pKa = 10.34SIEE358 pKa = 4.12KK359 pKa = 10.25DD360 pKa = 3.43GEE362 pKa = 4.13IQRR365 pKa = 11.84GCVWIPEE372 pKa = 4.39GQSGCQAVRR381 pKa = 11.84EE382 pKa = 4.3KK383 pKa = 11.25VDD385 pKa = 3.57LQQNSVNDD393 pKa = 3.96LCDD396 pKa = 3.03VCQTDD401 pKa = 3.3NCNGSASLRR410 pKa = 11.84MSLAALLLLLGLGLRR425 pKa = 11.84SLFF428 pKa = 3.96
MM1 pKa = 7.73RR2 pKa = 11.84LISLVIFCLCVCAAIWPANALQCYY26 pKa = 8.15QCIGSEE32 pKa = 4.5CDD34 pKa = 4.64DD35 pKa = 4.82LDD37 pKa = 3.94DD38 pKa = 5.46SKK40 pKa = 11.8VVSCNLDD47 pKa = 3.39EE48 pKa = 4.46TTSTLTTEE56 pKa = 4.24DD57 pKa = 3.63TTTEE61 pKa = 4.18SLPDD65 pKa = 3.49TSTTSSGSSTGGSTTSPGPTVDD87 pKa = 3.59TPATEE92 pKa = 4.21SSPPSTTDD100 pKa = 3.08QSDD103 pKa = 3.63TTTEE107 pKa = 4.08ATSEE111 pKa = 4.19TTTTGEE117 pKa = 4.02PSEE120 pKa = 4.33STEE123 pKa = 4.12TTEE126 pKa = 4.24TTTGEE131 pKa = 4.13PSGSTEE137 pKa = 4.2TTDD140 pKa = 3.14STTEE144 pKa = 3.95EE145 pKa = 4.29PSGSTEE151 pKa = 4.06TTEE154 pKa = 4.12STTEE158 pKa = 3.87STTDD162 pKa = 3.19STSDD166 pKa = 3.41STSDD170 pKa = 3.44STSDD174 pKa = 3.49STSDD178 pKa = 3.37SSPGSTASSTSEE190 pKa = 3.94STSEE194 pKa = 4.09STSDD198 pKa = 3.43STSDD202 pKa = 3.48STSDD206 pKa = 3.27SSSGSTAGSTSDD218 pKa = 3.41STSEE222 pKa = 4.09STSDD226 pKa = 3.42STSDD230 pKa = 3.19STTGSTSDD238 pKa = 3.48STSDD242 pKa = 3.44STSDD246 pKa = 3.38STSSPEE252 pKa = 3.7SSIPSSSGSTEE263 pKa = 3.86SSTAASTGSSEE274 pKa = 4.4TGEE277 pKa = 4.39STTSGSTEE285 pKa = 3.98SSTSSSASVVNPGNLEE301 pKa = 4.26NIQLALQQAALPGGIRR317 pKa = 11.84SGRR320 pKa = 11.84ARR322 pKa = 11.84RR323 pKa = 11.84SLAAQAADD331 pKa = 3.3QLIRR335 pKa = 11.84NYY337 pKa = 10.52RR338 pKa = 11.84SYY340 pKa = 11.16EE341 pKa = 3.86FNGRR345 pKa = 11.84ATNYY349 pKa = 7.59RR350 pKa = 11.84TTACYY355 pKa = 10.34SIEE358 pKa = 4.12KK359 pKa = 10.25DD360 pKa = 3.43GEE362 pKa = 4.13IQRR365 pKa = 11.84GCVWIPEE372 pKa = 4.39GQSGCQAVRR381 pKa = 11.84EE382 pKa = 4.3KK383 pKa = 11.25VDD385 pKa = 3.57LQQNSVNDD393 pKa = 3.96LCDD396 pKa = 3.03VCQTDD401 pKa = 3.3NCNGSASLRR410 pKa = 11.84MSLAALLLLLGLGLRR425 pKa = 11.84SLFF428 pKa = 3.96
Molecular weight: 43.57 kDa
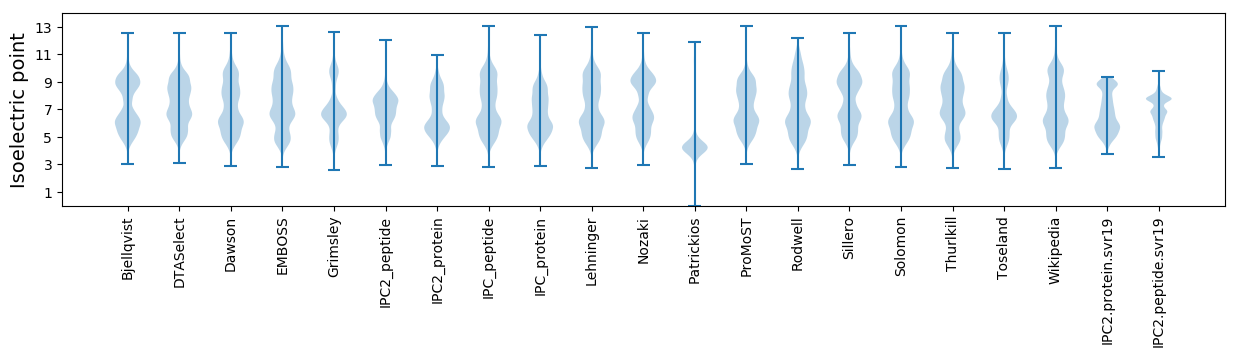
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B4LZV7|B4LZV7_DROVI Uncharacterized protein OS=Drosophila virilis OX=7244 GN=Dvir\GJ23227 PE=4 SV=2
MM1 pKa = 6.69VQTGISLNEE10 pKa = 4.84RR11 pKa = 11.84FTQMQSRR18 pKa = 11.84QQQPPAAARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SRR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84IVDD42 pKa = 3.85AAPSVANSRR51 pKa = 11.84LLQEE55 pKa = 5.38FKK57 pKa = 10.53RR58 pKa = 11.84KK59 pKa = 8.51HH60 pKa = 4.68TVQTALKK67 pKa = 9.27LKK69 pKa = 10.03RR70 pKa = 11.84RR71 pKa = 11.84SLRR74 pKa = 11.84TTAGGASARR83 pKa = 11.84LGGIKK88 pKa = 10.39ALRR91 pKa = 11.84LGANGKK97 pKa = 7.97PVRR100 pKa = 11.84SNNGTRR106 pKa = 11.84LATMRR111 pKa = 11.84ADD113 pKa = 5.64LIANNNNGRR122 pKa = 11.84ARR124 pKa = 11.84RR125 pKa = 11.84SSSSARR131 pKa = 11.84RR132 pKa = 11.84SSAAAGGLGQRR143 pKa = 11.84LGLRR147 pKa = 11.84RR148 pKa = 11.84PVVGGAAEE156 pKa = 3.94RR157 pKa = 11.84VAQRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84GLSRR167 pKa = 11.84QRR169 pKa = 11.84QQQQPLPQLQSRR181 pKa = 11.84GRR183 pKa = 11.84SRR185 pKa = 11.84SRR187 pKa = 11.84GRR189 pKa = 11.84ARR191 pKa = 11.84SLSRR195 pKa = 11.84GRR197 pKa = 11.84DD198 pKa = 3.32QSRR201 pKa = 11.84GRR203 pKa = 11.84SQSVGRR209 pKa = 11.84AQSRR213 pKa = 11.84GRR215 pKa = 11.84RR216 pKa = 11.84QLSANGGRR224 pKa = 11.84GVSAGGGGGRR234 pKa = 11.84APGRR238 pKa = 11.84VSVKK242 pKa = 9.43QRR244 pKa = 11.84LGVRR248 pKa = 11.84PGVAAAAAAAAKK260 pKa = 9.98GAANKK265 pKa = 9.95RR266 pKa = 11.84RR267 pKa = 11.84GNRR270 pKa = 11.84RR271 pKa = 11.84GNSQMRR277 pKa = 11.84GVTAGRR283 pKa = 11.84IDD285 pKa = 3.36KK286 pKa = 10.44RR287 pKa = 11.84RR288 pKa = 11.84NSSNTAQKK296 pKa = 8.76TVKK299 pKa = 10.07AAAGGRR305 pKa = 11.84GRR307 pKa = 11.84KK308 pKa = 8.93GRR310 pKa = 11.84ARR312 pKa = 11.84GRR314 pKa = 11.84SAVRR318 pKa = 11.84AAAAAVDD325 pKa = 3.7AASAATHH332 pKa = 6.76RR333 pKa = 11.84SRR335 pKa = 11.84SRR337 pKa = 11.84SRR339 pKa = 11.84SRR341 pKa = 11.84SRR343 pKa = 11.84ARR345 pKa = 11.84QGKK348 pKa = 8.32VAAGAAASSNGRR360 pKa = 11.84QKK362 pKa = 11.1GAGPVGARR370 pKa = 11.84RR371 pKa = 11.84RR372 pKa = 11.84GRR374 pKa = 11.84SLQRR378 pKa = 11.84TGVTVARR385 pKa = 11.84GANRR389 pKa = 11.84NGNKK393 pKa = 9.32KK394 pKa = 10.46KK395 pKa = 10.73NIITEE400 pKa = 4.35GVKK403 pKa = 10.27PNQLQQGRR411 pKa = 11.84RR412 pKa = 11.84GRR414 pKa = 11.84SRR416 pKa = 11.84GRR418 pKa = 11.84GGRR421 pKa = 11.84TVAGKK426 pKa = 7.53PQRR429 pKa = 11.84RR430 pKa = 11.84EE431 pKa = 3.91VKK433 pKa = 10.54RR434 pKa = 11.84EE435 pKa = 3.86DD436 pKa = 4.12LDD438 pKa = 4.45NEE440 pKa = 4.2LDD442 pKa = 3.56QYY444 pKa = 11.42MSTTKK449 pKa = 10.68SEE451 pKa = 3.92MDD453 pKa = 3.72FLLKK457 pKa = 10.68
MM1 pKa = 6.69VQTGISLNEE10 pKa = 4.84RR11 pKa = 11.84FTQMQSRR18 pKa = 11.84QQQPPAAARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SRR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84IVDD42 pKa = 3.85AAPSVANSRR51 pKa = 11.84LLQEE55 pKa = 5.38FKK57 pKa = 10.53RR58 pKa = 11.84KK59 pKa = 8.51HH60 pKa = 4.68TVQTALKK67 pKa = 9.27LKK69 pKa = 10.03RR70 pKa = 11.84RR71 pKa = 11.84SLRR74 pKa = 11.84TTAGGASARR83 pKa = 11.84LGGIKK88 pKa = 10.39ALRR91 pKa = 11.84LGANGKK97 pKa = 7.97PVRR100 pKa = 11.84SNNGTRR106 pKa = 11.84LATMRR111 pKa = 11.84ADD113 pKa = 5.64LIANNNNGRR122 pKa = 11.84ARR124 pKa = 11.84RR125 pKa = 11.84SSSSARR131 pKa = 11.84RR132 pKa = 11.84SSAAAGGLGQRR143 pKa = 11.84LGLRR147 pKa = 11.84RR148 pKa = 11.84PVVGGAAEE156 pKa = 3.94RR157 pKa = 11.84VAQRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84GLSRR167 pKa = 11.84QRR169 pKa = 11.84QQQQPLPQLQSRR181 pKa = 11.84GRR183 pKa = 11.84SRR185 pKa = 11.84SRR187 pKa = 11.84GRR189 pKa = 11.84ARR191 pKa = 11.84SLSRR195 pKa = 11.84GRR197 pKa = 11.84DD198 pKa = 3.32QSRR201 pKa = 11.84GRR203 pKa = 11.84SQSVGRR209 pKa = 11.84AQSRR213 pKa = 11.84GRR215 pKa = 11.84RR216 pKa = 11.84QLSANGGRR224 pKa = 11.84GVSAGGGGGRR234 pKa = 11.84APGRR238 pKa = 11.84VSVKK242 pKa = 9.43QRR244 pKa = 11.84LGVRR248 pKa = 11.84PGVAAAAAAAAKK260 pKa = 9.98GAANKK265 pKa = 9.95RR266 pKa = 11.84RR267 pKa = 11.84GNRR270 pKa = 11.84RR271 pKa = 11.84GNSQMRR277 pKa = 11.84GVTAGRR283 pKa = 11.84IDD285 pKa = 3.36KK286 pKa = 10.44RR287 pKa = 11.84RR288 pKa = 11.84NSSNTAQKK296 pKa = 8.76TVKK299 pKa = 10.07AAAGGRR305 pKa = 11.84GRR307 pKa = 11.84KK308 pKa = 8.93GRR310 pKa = 11.84ARR312 pKa = 11.84GRR314 pKa = 11.84SAVRR318 pKa = 11.84AAAAAVDD325 pKa = 3.7AASAATHH332 pKa = 6.76RR333 pKa = 11.84SRR335 pKa = 11.84SRR337 pKa = 11.84SRR339 pKa = 11.84SRR341 pKa = 11.84SRR343 pKa = 11.84ARR345 pKa = 11.84QGKK348 pKa = 8.32VAAGAAASSNGRR360 pKa = 11.84QKK362 pKa = 11.1GAGPVGARR370 pKa = 11.84RR371 pKa = 11.84RR372 pKa = 11.84GRR374 pKa = 11.84SLQRR378 pKa = 11.84TGVTVARR385 pKa = 11.84GANRR389 pKa = 11.84NGNKK393 pKa = 9.32KK394 pKa = 10.46KK395 pKa = 10.73NIITEE400 pKa = 4.35GVKK403 pKa = 10.27PNQLQQGRR411 pKa = 11.84RR412 pKa = 11.84GRR414 pKa = 11.84SRR416 pKa = 11.84GRR418 pKa = 11.84GGRR421 pKa = 11.84TVAGKK426 pKa = 7.53PQRR429 pKa = 11.84RR430 pKa = 11.84EE431 pKa = 3.91VKK433 pKa = 10.54RR434 pKa = 11.84EE435 pKa = 3.86DD436 pKa = 4.12LDD438 pKa = 4.45NEE440 pKa = 4.2LDD442 pKa = 3.56QYY444 pKa = 11.42MSTTKK449 pKa = 10.68SEE451 pKa = 3.92MDD453 pKa = 3.72FLLKK457 pKa = 10.68
Molecular weight: 48.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11705031 |
17 |
17765 |
639.1 |
71.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.904 ± 0.024 | 1.891 ± 0.024 |
5.178 ± 0.016 | 6.384 ± 0.032 |
3.381 ± 0.013 | 5.802 ± 0.026 |
2.621 ± 0.011 | 4.941 ± 0.017 |
5.518 ± 0.028 | 9.121 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.294 ± 0.011 | 4.893 ± 0.015 |
5.395 ± 0.024 | 5.55 ± 0.028 |
5.476 ± 0.014 | 8.093 ± 0.029 |
5.862 ± 0.016 | 5.713 ± 0.016 |
0.982 ± 0.006 | 2.998 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
