
Faeces associated gemycircularvirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus porci2; Porcine associated gemycircularvirus 2
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
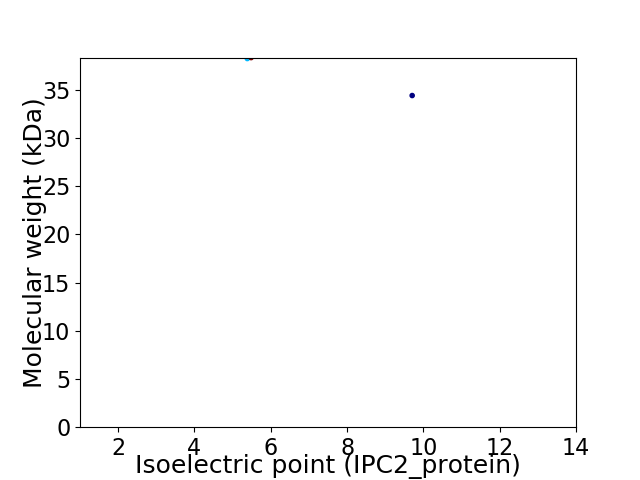
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
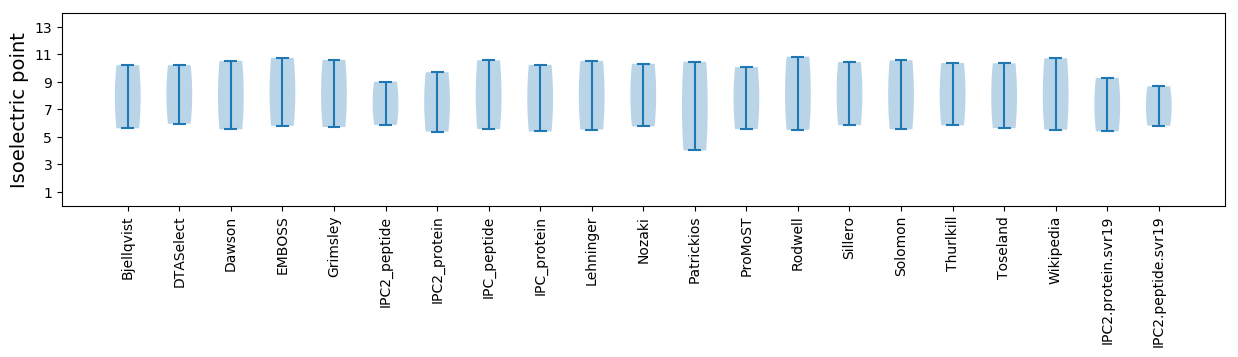
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YRY5|T1YRY5_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 2 OX=1391031 PE=3 SV=1
MM1 pKa = 7.59PPKK4 pKa = 9.78PAYY7 pKa = 9.28FVQSRR12 pKa = 11.84YY13 pKa = 10.12VLLTYY18 pKa = 9.15AQCGDD23 pKa = 3.48LDD25 pKa = 3.38AWAVNDD31 pKa = 3.99HH32 pKa = 6.7LSFLGAEE39 pKa = 4.43CIIGRR44 pKa = 11.84EE45 pKa = 4.04LHH47 pKa = 6.88ADD49 pKa = 3.84DD50 pKa = 4.86GTHH53 pKa = 6.64LHH55 pKa = 6.74AFCDD59 pKa = 5.35FSRR62 pKa = 11.84KK63 pKa = 8.65FRR65 pKa = 11.84SRR67 pKa = 11.84RR68 pKa = 11.84SDD70 pKa = 3.22VFDD73 pKa = 3.2VGGRR77 pKa = 11.84HH78 pKa = 6.08PNVMPSFGRR87 pKa = 11.84PEE89 pKa = 4.4GGWDD93 pKa = 3.39YY94 pKa = 10.83ATKK97 pKa = 10.84DD98 pKa = 3.25GDD100 pKa = 3.74VCAGGLPRR108 pKa = 11.84PSGRR112 pKa = 11.84GVYY115 pKa = 10.06EE116 pKa = 4.89DD117 pKa = 3.54VSSWSIIVSQEE128 pKa = 3.9SEE130 pKa = 4.43GEE132 pKa = 3.81FWEE135 pKa = 4.98CVARR139 pKa = 11.84LDD141 pKa = 5.14PKK143 pKa = 10.89ALCTNYY149 pKa = 10.53GSLRR153 pKa = 11.84AYY155 pKa = 10.43ANWKK159 pKa = 8.74YY160 pKa = 10.66RR161 pKa = 11.84PAPVQYY167 pKa = 9.46EE168 pKa = 4.24HH169 pKa = 7.25PAGIKK174 pKa = 10.2FEE176 pKa = 4.66LGNVPEE182 pKa = 4.19LVVWRR187 pKa = 11.84DD188 pKa = 2.78IALGVDD194 pKa = 3.7RR195 pKa = 11.84EE196 pKa = 4.11RR197 pKa = 11.84GILVIFGDD205 pKa = 3.67TRR207 pKa = 11.84LGKK210 pKa = 7.65TLWARR215 pKa = 11.84SIGPHH220 pKa = 6.89IYY222 pKa = 10.2TIGQMSGEE230 pKa = 4.15VLLRR234 pKa = 11.84DD235 pKa = 4.84GPDD238 pKa = 2.84ADD240 pKa = 3.79YY241 pKa = 11.53AVFDD245 pKa = 4.39DD246 pKa = 4.11MRR248 pKa = 11.84GGLEE252 pKa = 4.55FFHH255 pKa = 6.92GWKK258 pKa = 9.71EE259 pKa = 3.86WFGCQSVVTVKK270 pKa = 10.58KK271 pKa = 10.58LYY273 pKa = 9.92RR274 pKa = 11.84DD275 pKa = 3.77PVQMPWGKK283 pKa = 9.35PVVWLSNRR291 pKa = 11.84DD292 pKa = 3.4PRR294 pKa = 11.84EE295 pKa = 3.72EE296 pKa = 3.94LRR298 pKa = 11.84DD299 pKa = 4.12GITNHH304 pKa = 5.76TSMGKK309 pKa = 8.81QAAIEE314 pKa = 4.34GDD316 pKa = 3.96IKK318 pKa = 10.76WLDD321 pKa = 3.56GNCIFVEE328 pKa = 4.06LDD330 pKa = 3.15HH331 pKa = 7.64AIFRR335 pKa = 11.84ANTEE339 pKa = 3.84
MM1 pKa = 7.59PPKK4 pKa = 9.78PAYY7 pKa = 9.28FVQSRR12 pKa = 11.84YY13 pKa = 10.12VLLTYY18 pKa = 9.15AQCGDD23 pKa = 3.48LDD25 pKa = 3.38AWAVNDD31 pKa = 3.99HH32 pKa = 6.7LSFLGAEE39 pKa = 4.43CIIGRR44 pKa = 11.84EE45 pKa = 4.04LHH47 pKa = 6.88ADD49 pKa = 3.84DD50 pKa = 4.86GTHH53 pKa = 6.64LHH55 pKa = 6.74AFCDD59 pKa = 5.35FSRR62 pKa = 11.84KK63 pKa = 8.65FRR65 pKa = 11.84SRR67 pKa = 11.84RR68 pKa = 11.84SDD70 pKa = 3.22VFDD73 pKa = 3.2VGGRR77 pKa = 11.84HH78 pKa = 6.08PNVMPSFGRR87 pKa = 11.84PEE89 pKa = 4.4GGWDD93 pKa = 3.39YY94 pKa = 10.83ATKK97 pKa = 10.84DD98 pKa = 3.25GDD100 pKa = 3.74VCAGGLPRR108 pKa = 11.84PSGRR112 pKa = 11.84GVYY115 pKa = 10.06EE116 pKa = 4.89DD117 pKa = 3.54VSSWSIIVSQEE128 pKa = 3.9SEE130 pKa = 4.43GEE132 pKa = 3.81FWEE135 pKa = 4.98CVARR139 pKa = 11.84LDD141 pKa = 5.14PKK143 pKa = 10.89ALCTNYY149 pKa = 10.53GSLRR153 pKa = 11.84AYY155 pKa = 10.43ANWKK159 pKa = 8.74YY160 pKa = 10.66RR161 pKa = 11.84PAPVQYY167 pKa = 9.46EE168 pKa = 4.24HH169 pKa = 7.25PAGIKK174 pKa = 10.2FEE176 pKa = 4.66LGNVPEE182 pKa = 4.19LVVWRR187 pKa = 11.84DD188 pKa = 2.78IALGVDD194 pKa = 3.7RR195 pKa = 11.84EE196 pKa = 4.11RR197 pKa = 11.84GILVIFGDD205 pKa = 3.67TRR207 pKa = 11.84LGKK210 pKa = 7.65TLWARR215 pKa = 11.84SIGPHH220 pKa = 6.89IYY222 pKa = 10.2TIGQMSGEE230 pKa = 4.15VLLRR234 pKa = 11.84DD235 pKa = 4.84GPDD238 pKa = 2.84ADD240 pKa = 3.79YY241 pKa = 11.53AVFDD245 pKa = 4.39DD246 pKa = 4.11MRR248 pKa = 11.84GGLEE252 pKa = 4.55FFHH255 pKa = 6.92GWKK258 pKa = 9.71EE259 pKa = 3.86WFGCQSVVTVKK270 pKa = 10.58KK271 pKa = 10.58LYY273 pKa = 9.92RR274 pKa = 11.84DD275 pKa = 3.77PVQMPWGKK283 pKa = 9.35PVVWLSNRR291 pKa = 11.84DD292 pKa = 3.4PRR294 pKa = 11.84EE295 pKa = 3.72EE296 pKa = 3.94LRR298 pKa = 11.84DD299 pKa = 4.12GITNHH304 pKa = 5.76TSMGKK309 pKa = 8.81QAAIEE314 pKa = 4.34GDD316 pKa = 3.96IKK318 pKa = 10.76WLDD321 pKa = 3.56GNCIFVEE328 pKa = 4.06LDD330 pKa = 3.15HH331 pKa = 7.64AIFRR335 pKa = 11.84ANTEE339 pKa = 3.84
Molecular weight: 38.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YRY5|T1YRY5_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 2 OX=1391031 PE=3 SV=1
MM1 pKa = 7.77AYY3 pKa = 10.07ARR5 pKa = 11.84SRR7 pKa = 11.84AKK9 pKa = 10.54RR10 pKa = 11.84GTGRR14 pKa = 11.84SKK16 pKa = 10.79KK17 pKa = 10.14RR18 pKa = 11.84GGLRR22 pKa = 11.84PRR24 pKa = 11.84YY25 pKa = 8.65RR26 pKa = 11.84KK27 pKa = 9.56KK28 pKa = 10.31RR29 pKa = 11.84PAYY32 pKa = 8.77RR33 pKa = 11.84KK34 pKa = 9.66KK35 pKa = 10.77GAMTKK40 pKa = 10.33KK41 pKa = 10.33RR42 pKa = 11.84ILNLTSTKK50 pKa = 10.2KK51 pKa = 10.4RR52 pKa = 11.84NTMLQVANTSSVNGGPVTPGPGPYY76 pKa = 9.64VVSDD80 pKa = 3.84VNGSQYY86 pKa = 11.48SVFMPTAQDD95 pKa = 3.94LNDD98 pKa = 3.5NNGAVNTISNQAQRR112 pKa = 11.84TATTCFIKK120 pKa = 10.78GFNEE124 pKa = 4.54RR125 pKa = 11.84IRR127 pKa = 11.84IQTSSGIPWFWRR139 pKa = 11.84RR140 pKa = 11.84ICFRR144 pKa = 11.84ARR146 pKa = 11.84NTTFFLYY153 pKa = 9.59STTDD157 pKa = 3.43TPSSTTGPAYY167 pKa = 9.85TEE169 pKa = 4.38TSNGMQRR176 pKa = 11.84LYY178 pKa = 10.96MNLTINQSGQTIGNILGVLFRR199 pKa = 11.84GQTGIDD205 pKa = 2.99WVDD208 pKa = 3.38PQTASVDD215 pKa = 3.57TTRR218 pKa = 11.84VDD220 pKa = 3.57LVFDD224 pKa = 3.25KK225 pKa = 10.83RR226 pKa = 11.84YY227 pKa = 8.31TIKK230 pKa = 10.64SGNASGTVRR239 pKa = 11.84DD240 pKa = 3.78FNIWHH245 pKa = 7.34RR246 pKa = 11.84YY247 pKa = 7.27GKK249 pKa = 10.06NLVYY253 pKa = 10.79DD254 pKa = 4.24DD255 pKa = 5.35DD256 pKa = 5.22EE257 pKa = 5.66NGSTEE262 pKa = 3.66ATRR265 pKa = 11.84YY266 pKa = 10.47NSVNDD271 pKa = 3.42KK272 pKa = 11.02RR273 pKa = 11.84GGGDD277 pKa = 3.89LLIFDD282 pKa = 4.91IFTPGTGAGTADD294 pKa = 4.3LLQLTSTSTLYY305 pKa = 9.69WHH307 pKa = 7.1EE308 pKa = 4.2KK309 pKa = 9.32
MM1 pKa = 7.77AYY3 pKa = 10.07ARR5 pKa = 11.84SRR7 pKa = 11.84AKK9 pKa = 10.54RR10 pKa = 11.84GTGRR14 pKa = 11.84SKK16 pKa = 10.79KK17 pKa = 10.14RR18 pKa = 11.84GGLRR22 pKa = 11.84PRR24 pKa = 11.84YY25 pKa = 8.65RR26 pKa = 11.84KK27 pKa = 9.56KK28 pKa = 10.31RR29 pKa = 11.84PAYY32 pKa = 8.77RR33 pKa = 11.84KK34 pKa = 9.66KK35 pKa = 10.77GAMTKK40 pKa = 10.33KK41 pKa = 10.33RR42 pKa = 11.84ILNLTSTKK50 pKa = 10.2KK51 pKa = 10.4RR52 pKa = 11.84NTMLQVANTSSVNGGPVTPGPGPYY76 pKa = 9.64VVSDD80 pKa = 3.84VNGSQYY86 pKa = 11.48SVFMPTAQDD95 pKa = 3.94LNDD98 pKa = 3.5NNGAVNTISNQAQRR112 pKa = 11.84TATTCFIKK120 pKa = 10.78GFNEE124 pKa = 4.54RR125 pKa = 11.84IRR127 pKa = 11.84IQTSSGIPWFWRR139 pKa = 11.84RR140 pKa = 11.84ICFRR144 pKa = 11.84ARR146 pKa = 11.84NTTFFLYY153 pKa = 9.59STTDD157 pKa = 3.43TPSSTTGPAYY167 pKa = 9.85TEE169 pKa = 4.38TSNGMQRR176 pKa = 11.84LYY178 pKa = 10.96MNLTINQSGQTIGNILGVLFRR199 pKa = 11.84GQTGIDD205 pKa = 2.99WVDD208 pKa = 3.38PQTASVDD215 pKa = 3.57TTRR218 pKa = 11.84VDD220 pKa = 3.57LVFDD224 pKa = 3.25KK225 pKa = 10.83RR226 pKa = 11.84YY227 pKa = 8.31TIKK230 pKa = 10.64SGNASGTVRR239 pKa = 11.84DD240 pKa = 3.78FNIWHH245 pKa = 7.34RR246 pKa = 11.84YY247 pKa = 7.27GKK249 pKa = 10.06NLVYY253 pKa = 10.79DD254 pKa = 4.24DD255 pKa = 5.35DD256 pKa = 5.22EE257 pKa = 5.66NGSTEE262 pKa = 3.66ATRR265 pKa = 11.84YY266 pKa = 10.47NSVNDD271 pKa = 3.42KK272 pKa = 11.02RR273 pKa = 11.84GGGDD277 pKa = 3.89LLIFDD282 pKa = 4.91IFTPGTGAGTADD294 pKa = 4.3LLQLTSTSTLYY305 pKa = 9.69WHH307 pKa = 7.1EE308 pKa = 4.2KK309 pKa = 9.32
Molecular weight: 34.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
648 |
309 |
339 |
324.0 |
36.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.173 ± 0.431 | 1.543 ± 0.575 |
6.79 ± 0.827 | 3.858 ± 1.437 |
4.321 ± 0.281 | 10.031 ± 0.207 |
1.852 ± 0.773 | 4.784 ± 0.045 |
4.63 ± 0.559 | 6.636 ± 0.52 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.852 ± 0.058 | 4.784 ± 1.499 |
4.784 ± 0.578 | 3.086 ± 0.511 |
8.025 ± 0.458 | 6.327 ± 0.716 |
7.562 ± 3.038 | 6.481 ± 0.836 |
2.623 ± 0.645 | 3.858 ± 0.224 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
