
Tea plant line pattern virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus; unclassified Ilarvirus
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
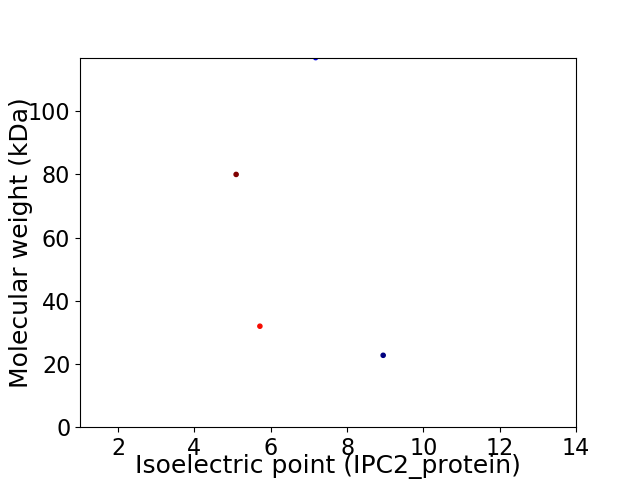
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A386QVH1|A0A386QVH1_9BROM Coat protein OS=Tea plant line pattern virus OX=2419940 PE=3 SV=1
MM1 pKa = 7.84DD2 pKa = 3.74VKK4 pKa = 10.89SWLSSLSSSSLDD16 pKa = 3.55PLRR19 pKa = 11.84EE20 pKa = 3.98ALLCDD25 pKa = 4.36DD26 pKa = 5.58DD27 pKa = 4.32MPIDD31 pKa = 3.59KK32 pKa = 10.98VEE34 pKa = 4.2EE35 pKa = 3.98NTHH38 pKa = 5.43ISRR41 pKa = 11.84FIGWVILRR49 pKa = 11.84KK50 pKa = 10.07VFFSEE55 pKa = 4.42EE56 pKa = 3.98FASDD60 pKa = 3.21EE61 pKa = 4.51YY62 pKa = 11.73YY63 pKa = 10.57FYY65 pKa = 11.35SDD67 pKa = 3.7CLRR70 pKa = 11.84LRR72 pKa = 11.84FFEE75 pKa = 4.42LFSVPMEE82 pKa = 4.7LYY84 pKa = 10.26DD85 pKa = 4.89ANDD88 pKa = 3.55SGSTNVVFVEE98 pKa = 4.67SDD100 pKa = 2.67ASYY103 pKa = 9.84YY104 pKa = 10.88KK105 pKa = 10.75EE106 pKa = 4.59MEE108 pKa = 4.55DD109 pKa = 4.33LDD111 pKa = 4.36LAFSFPSEE119 pKa = 4.3DD120 pKa = 3.68KK121 pKa = 11.12RR122 pKa = 11.84GDD124 pKa = 3.72VNDD127 pKa = 4.11SSPDD131 pKa = 3.56SVEE134 pKa = 3.88LTEE137 pKa = 4.2VSQVEE142 pKa = 4.19VEE144 pKa = 4.42PYY146 pKa = 10.89LDD148 pKa = 4.66FKK150 pKa = 11.76LEE152 pKa = 4.08VCTLPDD158 pKa = 4.34DD159 pKa = 4.95EE160 pKa = 4.84GLCRR164 pKa = 11.84QLDD167 pKa = 3.99WKK169 pKa = 10.69AQTHH173 pKa = 4.28VVVEE177 pKa = 4.14NVLRR181 pKa = 11.84NSSIEE186 pKa = 4.15IINSMRR192 pKa = 11.84TYY194 pKa = 10.66NANCIQDD201 pKa = 4.76AIDD204 pKa = 3.56EE205 pKa = 4.53VFPHH209 pKa = 6.66HH210 pKa = 6.87FSVDD214 pKa = 2.95DD215 pKa = 4.38RR216 pKa = 11.84YY217 pKa = 10.54FQTFVEE223 pKa = 4.61TSDD226 pKa = 3.79IDD228 pKa = 4.18LEE230 pKa = 4.63LDD232 pKa = 3.35AAHH235 pKa = 7.24IDD237 pKa = 3.43MSKK240 pKa = 10.33FPKK243 pKa = 10.47KK244 pKa = 10.3FDD246 pKa = 3.07IHH248 pKa = 6.96FDD250 pKa = 3.64WEE252 pKa = 4.59PKK254 pKa = 10.48LSTGLTSTRR263 pKa = 11.84YY264 pKa = 9.71NSFRR268 pKa = 11.84EE269 pKa = 3.64AALAIKK275 pKa = 9.8KK276 pKa = 10.33RR277 pKa = 11.84NLNVPDD283 pKa = 4.06LTADD287 pKa = 3.97LDD289 pKa = 3.98VDD291 pKa = 4.1SLVPKK296 pKa = 10.4VMEE299 pKa = 4.54RR300 pKa = 11.84FLKK303 pKa = 10.58ILDD306 pKa = 3.84LGKK309 pKa = 10.88LSGFPDD315 pKa = 4.21DD316 pKa = 4.38SHH318 pKa = 8.75DD319 pKa = 3.84VGCFLLDD326 pKa = 3.39YY327 pKa = 10.56LSRR330 pKa = 11.84KK331 pKa = 8.55GLSYY335 pKa = 11.24DD336 pKa = 3.54VLEE339 pKa = 4.78PGCMVSLNKK348 pKa = 9.26YY349 pKa = 7.76RR350 pKa = 11.84HH351 pKa = 5.72MIKK354 pKa = 10.08SQLKK358 pKa = 8.85PVEE361 pKa = 4.15EE362 pKa = 5.05DD363 pKa = 3.21SLHH366 pKa = 6.21VEE368 pKa = 4.27RR369 pKa = 11.84PLAATITYY377 pKa = 9.36HH378 pKa = 6.34GKK380 pKa = 10.1EE381 pKa = 4.29KK382 pKa = 11.08VSVTTPFFLSLAAKK396 pKa = 8.71LLCVLSDD403 pKa = 3.42KK404 pKa = 11.13VVIPMGKK411 pKa = 8.33EE412 pKa = 3.48HH413 pKa = 6.79QLFDD417 pKa = 3.84INVDD421 pKa = 3.37VFRR424 pKa = 11.84KK425 pKa = 9.19IKK427 pKa = 8.68FWKK430 pKa = 10.02EE431 pKa = 2.77IDD433 pKa = 3.36YY434 pKa = 11.51SKK436 pKa = 10.92FDD438 pKa = 3.38KK439 pKa = 11.22SQGKK443 pKa = 8.33LHH445 pKa = 6.86HH446 pKa = 6.86EE447 pKa = 3.78IQRR450 pKa = 11.84AIFRR454 pKa = 11.84DD455 pKa = 3.91LQCDD459 pKa = 3.44EE460 pKa = 5.22GFLSTWFHH468 pKa = 4.99SHH470 pKa = 5.81EE471 pKa = 4.39FSTIFDD477 pKa = 3.87SEE479 pKa = 4.27AGIGFSTDD487 pKa = 3.49FQRR490 pKa = 11.84RR491 pKa = 11.84TGDD494 pKa = 2.74ACTYY498 pKa = 10.46LGNTLVTLATLSFVYY513 pKa = 10.57DD514 pKa = 4.17LDD516 pKa = 4.55SPDD519 pKa = 3.06VGLIVVSGDD528 pKa = 3.37DD529 pKa = 3.51SLIGTFHH536 pKa = 7.07SEE538 pKa = 3.92LPRR541 pKa = 11.84EE542 pKa = 3.98RR543 pKa = 11.84EE544 pKa = 4.09ILCSTLFNFEE554 pKa = 3.96TKK556 pKa = 10.16FPHH559 pKa = 5.6NQPFICSKK567 pKa = 10.23FLVEE571 pKa = 5.95IEE573 pKa = 4.34TTDD576 pKa = 3.24GRR578 pKa = 11.84GKK580 pKa = 10.33VVAVPSPIKK589 pKa = 10.64LLMKK593 pKa = 10.51LGPKK597 pKa = 10.07SMNKK601 pKa = 9.25QLFPAWVEE609 pKa = 4.25SIKK612 pKa = 10.95DD613 pKa = 3.34YY614 pKa = 11.13AAPFRR619 pKa = 11.84NAFVAEE625 pKa = 4.22QVCNLASYY633 pKa = 10.79RR634 pKa = 11.84FGRR637 pKa = 11.84RR638 pKa = 11.84CFTFLLPAISEE649 pKa = 4.44VLWMLSNPNVKK660 pKa = 10.27LKK662 pKa = 10.92EE663 pKa = 3.94KK664 pKa = 10.82LFLQNSSYY672 pKa = 11.09LSNHH676 pKa = 4.96CHH678 pKa = 7.04DD679 pKa = 3.93RR680 pKa = 11.84RR681 pKa = 11.84EE682 pKa = 4.2KK683 pKa = 9.75EE684 pKa = 4.03KK685 pKa = 10.87KK686 pKa = 9.41RR687 pKa = 11.84NRR689 pKa = 11.84PRR691 pKa = 11.84ARR693 pKa = 11.84SKK695 pKa = 10.78RR696 pKa = 3.69
MM1 pKa = 7.84DD2 pKa = 3.74VKK4 pKa = 10.89SWLSSLSSSSLDD16 pKa = 3.55PLRR19 pKa = 11.84EE20 pKa = 3.98ALLCDD25 pKa = 4.36DD26 pKa = 5.58DD27 pKa = 4.32MPIDD31 pKa = 3.59KK32 pKa = 10.98VEE34 pKa = 4.2EE35 pKa = 3.98NTHH38 pKa = 5.43ISRR41 pKa = 11.84FIGWVILRR49 pKa = 11.84KK50 pKa = 10.07VFFSEE55 pKa = 4.42EE56 pKa = 3.98FASDD60 pKa = 3.21EE61 pKa = 4.51YY62 pKa = 11.73YY63 pKa = 10.57FYY65 pKa = 11.35SDD67 pKa = 3.7CLRR70 pKa = 11.84LRR72 pKa = 11.84FFEE75 pKa = 4.42LFSVPMEE82 pKa = 4.7LYY84 pKa = 10.26DD85 pKa = 4.89ANDD88 pKa = 3.55SGSTNVVFVEE98 pKa = 4.67SDD100 pKa = 2.67ASYY103 pKa = 9.84YY104 pKa = 10.88KK105 pKa = 10.75EE106 pKa = 4.59MEE108 pKa = 4.55DD109 pKa = 4.33LDD111 pKa = 4.36LAFSFPSEE119 pKa = 4.3DD120 pKa = 3.68KK121 pKa = 11.12RR122 pKa = 11.84GDD124 pKa = 3.72VNDD127 pKa = 4.11SSPDD131 pKa = 3.56SVEE134 pKa = 3.88LTEE137 pKa = 4.2VSQVEE142 pKa = 4.19VEE144 pKa = 4.42PYY146 pKa = 10.89LDD148 pKa = 4.66FKK150 pKa = 11.76LEE152 pKa = 4.08VCTLPDD158 pKa = 4.34DD159 pKa = 4.95EE160 pKa = 4.84GLCRR164 pKa = 11.84QLDD167 pKa = 3.99WKK169 pKa = 10.69AQTHH173 pKa = 4.28VVVEE177 pKa = 4.14NVLRR181 pKa = 11.84NSSIEE186 pKa = 4.15IINSMRR192 pKa = 11.84TYY194 pKa = 10.66NANCIQDD201 pKa = 4.76AIDD204 pKa = 3.56EE205 pKa = 4.53VFPHH209 pKa = 6.66HH210 pKa = 6.87FSVDD214 pKa = 2.95DD215 pKa = 4.38RR216 pKa = 11.84YY217 pKa = 10.54FQTFVEE223 pKa = 4.61TSDD226 pKa = 3.79IDD228 pKa = 4.18LEE230 pKa = 4.63LDD232 pKa = 3.35AAHH235 pKa = 7.24IDD237 pKa = 3.43MSKK240 pKa = 10.33FPKK243 pKa = 10.47KK244 pKa = 10.3FDD246 pKa = 3.07IHH248 pKa = 6.96FDD250 pKa = 3.64WEE252 pKa = 4.59PKK254 pKa = 10.48LSTGLTSTRR263 pKa = 11.84YY264 pKa = 9.71NSFRR268 pKa = 11.84EE269 pKa = 3.64AALAIKK275 pKa = 9.8KK276 pKa = 10.33RR277 pKa = 11.84NLNVPDD283 pKa = 4.06LTADD287 pKa = 3.97LDD289 pKa = 3.98VDD291 pKa = 4.1SLVPKK296 pKa = 10.4VMEE299 pKa = 4.54RR300 pKa = 11.84FLKK303 pKa = 10.58ILDD306 pKa = 3.84LGKK309 pKa = 10.88LSGFPDD315 pKa = 4.21DD316 pKa = 4.38SHH318 pKa = 8.75DD319 pKa = 3.84VGCFLLDD326 pKa = 3.39YY327 pKa = 10.56LSRR330 pKa = 11.84KK331 pKa = 8.55GLSYY335 pKa = 11.24DD336 pKa = 3.54VLEE339 pKa = 4.78PGCMVSLNKK348 pKa = 9.26YY349 pKa = 7.76RR350 pKa = 11.84HH351 pKa = 5.72MIKK354 pKa = 10.08SQLKK358 pKa = 8.85PVEE361 pKa = 4.15EE362 pKa = 5.05DD363 pKa = 3.21SLHH366 pKa = 6.21VEE368 pKa = 4.27RR369 pKa = 11.84PLAATITYY377 pKa = 9.36HH378 pKa = 6.34GKK380 pKa = 10.1EE381 pKa = 4.29KK382 pKa = 11.08VSVTTPFFLSLAAKK396 pKa = 8.71LLCVLSDD403 pKa = 3.42KK404 pKa = 11.13VVIPMGKK411 pKa = 8.33EE412 pKa = 3.48HH413 pKa = 6.79QLFDD417 pKa = 3.84INVDD421 pKa = 3.37VFRR424 pKa = 11.84KK425 pKa = 9.19IKK427 pKa = 8.68FWKK430 pKa = 10.02EE431 pKa = 2.77IDD433 pKa = 3.36YY434 pKa = 11.51SKK436 pKa = 10.92FDD438 pKa = 3.38KK439 pKa = 11.22SQGKK443 pKa = 8.33LHH445 pKa = 6.86HH446 pKa = 6.86EE447 pKa = 3.78IQRR450 pKa = 11.84AIFRR454 pKa = 11.84DD455 pKa = 3.91LQCDD459 pKa = 3.44EE460 pKa = 5.22GFLSTWFHH468 pKa = 4.99SHH470 pKa = 5.81EE471 pKa = 4.39FSTIFDD477 pKa = 3.87SEE479 pKa = 4.27AGIGFSTDD487 pKa = 3.49FQRR490 pKa = 11.84RR491 pKa = 11.84TGDD494 pKa = 2.74ACTYY498 pKa = 10.46LGNTLVTLATLSFVYY513 pKa = 10.57DD514 pKa = 4.17LDD516 pKa = 4.55SPDD519 pKa = 3.06VGLIVVSGDD528 pKa = 3.37DD529 pKa = 3.51SLIGTFHH536 pKa = 7.07SEE538 pKa = 3.92LPRR541 pKa = 11.84EE542 pKa = 3.98RR543 pKa = 11.84EE544 pKa = 4.09ILCSTLFNFEE554 pKa = 3.96TKK556 pKa = 10.16FPHH559 pKa = 5.6NQPFICSKK567 pKa = 10.23FLVEE571 pKa = 5.95IEE573 pKa = 4.34TTDD576 pKa = 3.24GRR578 pKa = 11.84GKK580 pKa = 10.33VVAVPSPIKK589 pKa = 10.64LLMKK593 pKa = 10.51LGPKK597 pKa = 10.07SMNKK601 pKa = 9.25QLFPAWVEE609 pKa = 4.25SIKK612 pKa = 10.95DD613 pKa = 3.34YY614 pKa = 11.13AAPFRR619 pKa = 11.84NAFVAEE625 pKa = 4.22QVCNLASYY633 pKa = 10.79RR634 pKa = 11.84FGRR637 pKa = 11.84RR638 pKa = 11.84CFTFLLPAISEE649 pKa = 4.44VLWMLSNPNVKK660 pKa = 10.27LKK662 pKa = 10.92EE663 pKa = 3.94KK664 pKa = 10.82LFLQNSSYY672 pKa = 11.09LSNHH676 pKa = 4.96CHH678 pKa = 7.04DD679 pKa = 3.93RR680 pKa = 11.84RR681 pKa = 11.84EE682 pKa = 4.2KK683 pKa = 9.75EE684 pKa = 4.03KK685 pKa = 10.87KK686 pKa = 9.41RR687 pKa = 11.84NRR689 pKa = 11.84PRR691 pKa = 11.84ARR693 pKa = 11.84SKK695 pKa = 10.78RR696 pKa = 3.69
Molecular weight: 80.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A386QVU9|A0A386QVU9_9BROM Movement protein OS=Tea plant line pattern virus OX=2419940 PE=4 SV=1
MM1 pKa = 7.13STNTQQKK8 pKa = 8.25KK9 pKa = 6.54TRR11 pKa = 11.84QNKK14 pKa = 9.44RR15 pKa = 11.84STQFGQNRR23 pKa = 11.84AKK25 pKa = 10.32QRR27 pKa = 11.84AQSTGEE33 pKa = 3.96SSTVPNFGTLRR44 pKa = 11.84LEE46 pKa = 4.16WVRR49 pKa = 11.84RR50 pKa = 11.84GPGDD54 pKa = 4.42LDD56 pKa = 3.15ILQFPYY62 pKa = 10.43QWVAKK67 pKa = 10.37AEE69 pKa = 4.21SSLKK73 pKa = 9.72ATSNDD78 pKa = 1.88IWYY81 pKa = 9.62IIDD84 pKa = 3.16LHH86 pKa = 6.75EE87 pKa = 4.18YY88 pKa = 8.7VKK90 pKa = 11.41DD91 pKa = 3.39MMEE94 pKa = 4.4FPTSVKK100 pKa = 10.24GFVFLFEE107 pKa = 5.36ANLSGQACLVTKK119 pKa = 9.54KK120 pKa = 9.21TGSFKK125 pKa = 11.05DD126 pKa = 3.44AFSSLNAFKK135 pKa = 10.72FEE137 pKa = 4.55KK138 pKa = 10.27GCHH141 pKa = 4.82TAVQLLAPSILTFATFSAGEE161 pKa = 3.9VALVFKK167 pKa = 11.04FEE169 pKa = 4.18GTLTAGAEE177 pKa = 4.02FMTRR181 pKa = 11.84KK182 pKa = 9.56VWGQSSMLPKK192 pKa = 10.47VEE194 pKa = 3.99INKK197 pKa = 9.53NLLRR201 pKa = 11.84GQQ203 pKa = 3.52
MM1 pKa = 7.13STNTQQKK8 pKa = 8.25KK9 pKa = 6.54TRR11 pKa = 11.84QNKK14 pKa = 9.44RR15 pKa = 11.84STQFGQNRR23 pKa = 11.84AKK25 pKa = 10.32QRR27 pKa = 11.84AQSTGEE33 pKa = 3.96SSTVPNFGTLRR44 pKa = 11.84LEE46 pKa = 4.16WVRR49 pKa = 11.84RR50 pKa = 11.84GPGDD54 pKa = 4.42LDD56 pKa = 3.15ILQFPYY62 pKa = 10.43QWVAKK67 pKa = 10.37AEE69 pKa = 4.21SSLKK73 pKa = 9.72ATSNDD78 pKa = 1.88IWYY81 pKa = 9.62IIDD84 pKa = 3.16LHH86 pKa = 6.75EE87 pKa = 4.18YY88 pKa = 8.7VKK90 pKa = 11.41DD91 pKa = 3.39MMEE94 pKa = 4.4FPTSVKK100 pKa = 10.24GFVFLFEE107 pKa = 5.36ANLSGQACLVTKK119 pKa = 9.54KK120 pKa = 9.21TGSFKK125 pKa = 11.05DD126 pKa = 3.44AFSSLNAFKK135 pKa = 10.72FEE137 pKa = 4.55KK138 pKa = 10.27GCHH141 pKa = 4.82TAVQLLAPSILTFATFSAGEE161 pKa = 3.9VALVFKK167 pKa = 11.04FEE169 pKa = 4.18GTLTAGAEE177 pKa = 4.02FMTRR181 pKa = 11.84KK182 pKa = 9.56VWGQSSMLPKK192 pKa = 10.47VEE194 pKa = 3.99INKK197 pKa = 9.53NLLRR201 pKa = 11.84GQQ203 pKa = 3.52
Molecular weight: 22.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2230 |
203 |
1042 |
557.5 |
62.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.65 ± 0.533 | 2.197 ± 0.255 |
6.413 ± 0.937 | 6.233 ± 0.419 |
5.381 ± 0.931 | 4.529 ± 0.496 |
2.556 ± 0.418 | 5.336 ± 0.605 |
6.637 ± 0.329 | 9.507 ± 0.506 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.511 ± 0.276 | 4.619 ± 0.549 |
3.812 ± 0.219 | 3.004 ± 0.647 |
5.291 ± 0.2 | 9.552 ± 0.437 |
5.919 ± 0.665 | 7.489 ± 0.493 |
0.942 ± 0.231 | 2.422 ± 0.248 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
