
Gelidibacter gilvus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Gelidibacter
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
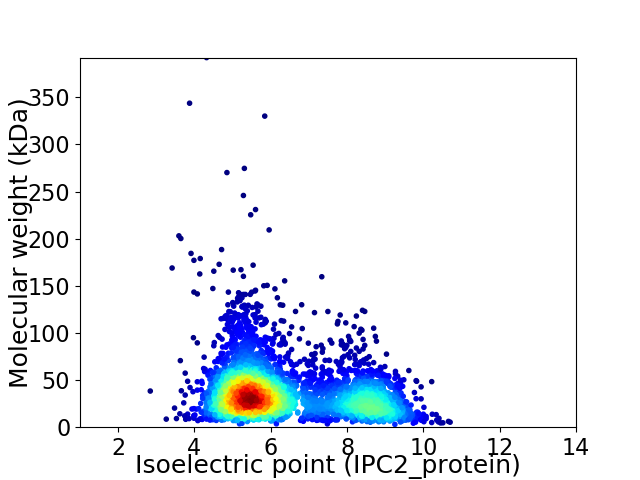
Virtual 2D-PAGE plot for 3732 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q0XA07|A0A4Q0XA07_9FLAO IS1182 family transposase OS=Gelidibacter gilvus OX=59602 GN=ESZ48_19115 PE=4 SV=1
AA1 pKa = 8.02QVITQDD7 pKa = 3.63ISIDD11 pKa = 3.36LDD13 pKa = 3.95ANGDD17 pKa = 3.54ASIIAAQVDD26 pKa = 3.79NGSNDD31 pKa = 3.12ACGIASMTVVPNTFSCSNIGANTVTLTVVDD61 pKa = 3.97NNGNSSSATATVTVNDD77 pKa = 3.41VTVAQVITQDD87 pKa = 3.24ISIDD91 pKa = 3.36LDD93 pKa = 3.95ANGDD97 pKa = 3.54ASIIAAQVDD106 pKa = 3.79NGSNDD111 pKa = 3.12ACGIASMTVVPSTFDD126 pKa = 3.22CSNIGTNTVTLTVVDD141 pKa = 3.97NNGNSSSATATVTVNDD157 pKa = 3.3VTAAQVITQDD167 pKa = 3.52ISIDD171 pKa = 3.36LDD173 pKa = 3.95ANGDD177 pKa = 3.54ASIIAAQVDD186 pKa = 3.79NGSNDD191 pKa = 3.12ACGIASMTVVPNTFDD206 pKa = 3.67CSNIGANTVTLTVVDD221 pKa = 4.02VNGNSSSATATVTVNDD237 pKa = 3.36VTLAQVITQDD247 pKa = 3.68ISIDD251 pKa = 3.36LDD253 pKa = 3.95ANGDD257 pKa = 3.54ASIIAAQVDD266 pKa = 3.79NGSNDD271 pKa = 3.12ACGIASMTVVPNTFDD286 pKa = 3.67CSNIGANTVTLTVVDD301 pKa = 3.97NNGNSSSATATVTVNDD317 pKa = 3.42VTVAQVVTQDD327 pKa = 3.2ISIDD331 pKa = 3.54LDD333 pKa = 3.6ASGDD337 pKa = 3.58ASIIAAQVDD346 pKa = 4.1DD347 pKa = 4.94GSNDD351 pKa = 3.14ACGIASMTVVPNTFDD366 pKa = 3.67CSNIGANTVTLTVVDD381 pKa = 4.02VNGNSSSATATVTVNDD397 pKa = 3.3VTAAQVITQDD407 pKa = 3.52ISIDD411 pKa = 3.36LDD413 pKa = 3.95ANGDD417 pKa = 3.54ASIIAAQVDD426 pKa = 3.79NGSNDD431 pKa = 3.12ACGIASMTVVPNTFDD446 pKa = 3.67CSNIGANTVTLTVVDD461 pKa = 3.97NNGNSSSATATVTVNDD477 pKa = 3.3VTAAQVITQNTSIDD491 pKa = 3.4LDD493 pKa = 4.08ANGNASIVPADD504 pKa = 3.8VDD506 pKa = 3.65NGSNDD511 pKa = 3.17ACGIASMTVTPNTFDD526 pKa = 3.9CSNIGANTVTLTVVDD541 pKa = 4.76LNGNSNSAMATVTVNDD557 pKa = 3.93VIAAQVITQNISLDD571 pKa = 3.42LDD573 pKa = 4.15ANGGASIVPADD584 pKa = 3.9VDD586 pKa = 3.79NGSSDD591 pKa = 3.37ACGIASMTVTPNTFDD606 pKa = 3.7CSNVGANTVTLTVTDD621 pKa = 3.47VNGNVSSATANVTVNDD637 pKa = 3.63ITPAIVITQNTSIDD651 pKa = 3.4LDD653 pKa = 4.08ANGNASIVPADD664 pKa = 3.8VDD666 pKa = 3.65NGSNDD671 pKa = 3.04ACGIANMTVSPNTFDD686 pKa = 4.69CSNLGMNTVTLTVTDD701 pKa = 3.66VNGNISSATAVVSISDD717 pKa = 3.48NTAPDD722 pKa = 3.93IVCVPDD728 pKa = 3.5GTRR731 pKa = 11.84DD732 pKa = 3.56TDD734 pKa = 3.91PGQCTYY740 pKa = 10.04TIQGAEE746 pKa = 3.89FDD748 pKa = 5.84AIFTDD753 pKa = 3.86NCSSGVITNSINGTATLAGEE773 pKa = 4.58VFMQGATEE781 pKa = 4.21VTWIVDD787 pKa = 3.59DD788 pKa = 4.21GHH790 pKa = 6.55GQTASCTTTITVEE803 pKa = 4.14DD804 pKa = 4.04NEE806 pKa = 4.43APIVDD811 pKa = 5.13CINIQVLLDD820 pKa = 3.46ASGNGTITVADD831 pKa = 3.82INNNSTDD838 pKa = 3.13NCGIASITLSQTSFDD853 pKa = 4.26CSDD856 pKa = 2.98IGGDD860 pKa = 3.74LDD862 pKa = 3.71EE863 pKa = 6.1LIISEE868 pKa = 4.29YY869 pKa = 11.04SDD871 pKa = 3.13GTGNNDD877 pKa = 4.48CIEE880 pKa = 4.37IYY882 pKa = 10.71NGTGNPINLYY892 pKa = 10.8AEE894 pKa = 5.54GYY896 pKa = 6.78TLRR899 pKa = 11.84FYY901 pKa = 11.72YY902 pKa = 10.49NGSTSYY908 pKa = 9.54TQVPLLGTVADD919 pKa = 3.98RR920 pKa = 11.84EE921 pKa = 4.72VYY923 pKa = 9.54VVCNPFGPGAMQADD937 pKa = 3.7QTGNFGFDD945 pKa = 3.27GNDD948 pKa = 3.47AIALTKK954 pKa = 10.45AGNAIDD960 pKa = 4.31VIGQIGVNPGTGWTVGSNSTAGTTLVRR987 pKa = 11.84NKK989 pKa = 10.56DD990 pKa = 3.51VLYY993 pKa = 11.3GNINDD998 pKa = 3.49ISGISSEE1005 pKa = 3.94WTQYY1009 pKa = 9.75AQNNTSNLGSHH1020 pKa = 6.87EE1021 pKa = 4.1IEE1023 pKa = 4.38IADD1026 pKa = 3.86LAKK1029 pKa = 10.79NVILTVTDD1037 pKa = 3.2TSGNVSTCEE1046 pKa = 3.96GNVTVIDD1053 pKa = 4.45NIAPVAMCQSVTIQLDD1069 pKa = 3.72ANGVASVTASEE1080 pKa = 4.45VDD1082 pKa = 3.22NGSNDD1087 pKa = 3.16ACGIASLVLDD1097 pKa = 3.75QTDD1100 pKa = 4.32FTCANLGDD1108 pKa = 3.86NTVILTVTDD1117 pKa = 3.61NNGNSSSCEE1126 pKa = 3.62ATITVEE1132 pKa = 4.75DD1133 pKa = 4.69NIDD1136 pKa = 3.82PTVMTQDD1143 pKa = 3.64LTIQLDD1149 pKa = 4.07ANGSASITPADD1160 pKa = 4.14IDD1162 pKa = 3.69NGSIDD1167 pKa = 3.49NCGIATQTVTPNSFDD1182 pKa = 3.59CSNVGVNTVTLTVTDD1197 pKa = 3.44TSGNVSTGIATVTVEE1212 pKa = 5.14DD1213 pKa = 3.93NVAPIAICKK1222 pKa = 10.16NITVEE1227 pKa = 4.28LGINGEE1233 pKa = 4.13ATITGADD1240 pKa = 3.56VDD1242 pKa = 4.41NGSSDD1247 pKa = 3.35ACGIASLDD1255 pKa = 3.4VSPSVFGCADD1265 pKa = 3.02IGANTVTLTVTDD1277 pKa = 3.61NNGNVSTCSATVTVTGIVPVVTISQGPLPEE1307 pKa = 4.45FCQGAVLVLTAEE1319 pKa = 4.09SDD1321 pKa = 3.44EE1322 pKa = 4.41DD1323 pKa = 5.2FGYY1326 pKa = 11.06LWTTGEE1332 pKa = 4.34TTQSIEE1338 pKa = 3.71IPGNGTYY1345 pKa = 10.78GVTVTSLTNCSTEE1358 pKa = 3.97AEE1360 pKa = 4.08ITITGFNAGVLVSSYY1375 pKa = 10.74TIIASEE1381 pKa = 4.28KK1382 pKa = 10.43VEE1384 pKa = 4.19LKK1386 pKa = 11.13NNVIVQTGGVGVTKK1400 pKa = 10.68ASGEE1404 pKa = 4.08IKK1406 pKa = 9.92IEE1408 pKa = 4.05KK1409 pKa = 9.54ASHH1412 pKa = 5.5IVGLGQANKK1421 pKa = 10.13IEE1423 pKa = 4.28IKK1425 pKa = 10.28QGSSVGTAVYY1435 pKa = 10.33QPANPTIPAFVYY1447 pKa = 10.26NGYY1450 pKa = 10.08SSSSSPDD1457 pKa = 2.81VRR1459 pKa = 11.84VNNNQTLTLSGGVYY1473 pKa = 10.46NKK1475 pKa = 10.06VEE1477 pKa = 4.23LEE1479 pKa = 4.08EE1480 pKa = 4.19NATVLFTASNVYY1492 pKa = 10.3INEE1495 pKa = 4.17LKK1497 pKa = 9.52TKK1499 pKa = 10.22KK1500 pKa = 10.33SATIEE1505 pKa = 4.12FNGCTNVFLNKK1516 pKa = 9.67EE1517 pKa = 4.22FKK1519 pKa = 10.89LDD1521 pKa = 3.79DD1522 pKa = 3.97NGVFNSNGYY1531 pKa = 8.33MVTVYY1536 pKa = 10.59INDD1539 pKa = 3.52DD1540 pKa = 3.87FEE1542 pKa = 4.69VKK1544 pKa = 10.25KK1545 pKa = 11.2GSDD1548 pKa = 3.23FTGRR1552 pKa = 11.84VHH1554 pKa = 6.79TNNHH1558 pKa = 6.24DD1559 pKa = 3.7IEE1561 pKa = 4.83VEE1563 pKa = 4.11GHH1565 pKa = 5.97NSSTTYY1571 pKa = 7.51MTGLFIGKK1579 pKa = 9.26KK1580 pKa = 9.73VEE1582 pKa = 3.81ADD1584 pKa = 3.84KK1585 pKa = 11.54NVVWNADD1592 pKa = 3.95QYY1594 pKa = 11.69CDD1596 pKa = 3.93PCPIEE1601 pKa = 5.2APINNGGSEE1610 pKa = 4.8PITDD1614 pKa = 3.53YY1615 pKa = 11.25TIIAFDD1621 pKa = 3.88EE1622 pKa = 4.46VHH1624 pKa = 6.27LHH1626 pKa = 6.69GDD1628 pKa = 3.73NIVQTGGVGVTRR1640 pKa = 11.84HH1641 pKa = 5.19KK1642 pKa = 11.0KK1643 pKa = 9.89KK1644 pKa = 10.53IKK1646 pKa = 9.35LHH1648 pKa = 6.22KK1649 pKa = 9.85DD1650 pKa = 2.95SHH1652 pKa = 5.22ITEE1655 pKa = 4.38FAKK1658 pKa = 10.7ASQIQVNGGSSIGTRR1673 pKa = 11.84ILSPADD1679 pKa = 3.82PIIPLFVKK1687 pKa = 10.38NVYY1690 pKa = 10.51SNSSSPNATINSGQTVTLTGSVYY1713 pKa = 11.08DD1714 pKa = 4.39KK1715 pKa = 11.58VDD1717 pKa = 3.72LKK1719 pKa = 11.24EE1720 pKa = 3.97GATVIFAQSNVFINEE1735 pKa = 3.91LKK1737 pKa = 7.7TRR1739 pKa = 11.84KK1740 pKa = 9.39YY1741 pKa = 10.22ATIKK1745 pKa = 10.71FSGCTNVFIKK1755 pKa = 10.77KK1756 pKa = 8.76EE1757 pKa = 4.14FKK1759 pKa = 10.61LDD1761 pKa = 3.62DD1762 pKa = 3.85YY1763 pKa = 11.89GVINPEE1769 pKa = 3.86TDD1771 pKa = 2.59GHH1773 pKa = 5.84KK1774 pKa = 10.7VVFHH1778 pKa = 6.73VDD1780 pKa = 3.25DD1781 pKa = 4.75DD1782 pKa = 4.58VEE1784 pKa = 4.12IDD1786 pKa = 3.31KK1787 pKa = 11.39GSVVVASIYY1796 pKa = 10.69AYY1798 pKa = 10.64NDD1800 pKa = 3.41EE1801 pKa = 4.41IKK1803 pKa = 10.76VDD1805 pKa = 4.09GSSSDD1810 pKa = 3.68PVSMKK1815 pKa = 10.87GLFIAKK1821 pKa = 9.21KK1822 pKa = 8.11VHH1824 pKa = 5.15AHH1826 pKa = 7.15EE1827 pKa = 4.85DD1828 pKa = 3.75VLWNKK1833 pKa = 8.19DD1834 pKa = 3.45TSGSPCPVITPTAQAPVAPEE1854 pKa = 3.6VGNRR1858 pKa = 11.84SIEE1861 pKa = 4.39LISVKK1866 pKa = 10.18AWPNPSDD1873 pKa = 3.44TVFNLRR1879 pKa = 11.84LITEE1883 pKa = 4.13NQVDD1887 pKa = 4.25VVSILVFDD1895 pKa = 4.49SNNKK1899 pKa = 9.0LVHH1902 pKa = 6.35SSTFRR1907 pKa = 11.84PEE1909 pKa = 3.46AVYY1912 pKa = 10.61QFGNEE1917 pKa = 4.11LEE1919 pKa = 4.24SGVYY1923 pKa = 8.69IVKK1926 pKa = 10.19VSQAGSNAYY1935 pKa = 8.71TRR1937 pKa = 11.84VIKK1940 pKa = 10.41FF1941 pKa = 3.24
AA1 pKa = 8.02QVITQDD7 pKa = 3.63ISIDD11 pKa = 3.36LDD13 pKa = 3.95ANGDD17 pKa = 3.54ASIIAAQVDD26 pKa = 3.79NGSNDD31 pKa = 3.12ACGIASMTVVPNTFSCSNIGANTVTLTVVDD61 pKa = 3.97NNGNSSSATATVTVNDD77 pKa = 3.41VTVAQVITQDD87 pKa = 3.24ISIDD91 pKa = 3.36LDD93 pKa = 3.95ANGDD97 pKa = 3.54ASIIAAQVDD106 pKa = 3.79NGSNDD111 pKa = 3.12ACGIASMTVVPSTFDD126 pKa = 3.22CSNIGTNTVTLTVVDD141 pKa = 3.97NNGNSSSATATVTVNDD157 pKa = 3.3VTAAQVITQDD167 pKa = 3.52ISIDD171 pKa = 3.36LDD173 pKa = 3.95ANGDD177 pKa = 3.54ASIIAAQVDD186 pKa = 3.79NGSNDD191 pKa = 3.12ACGIASMTVVPNTFDD206 pKa = 3.67CSNIGANTVTLTVVDD221 pKa = 4.02VNGNSSSATATVTVNDD237 pKa = 3.36VTLAQVITQDD247 pKa = 3.68ISIDD251 pKa = 3.36LDD253 pKa = 3.95ANGDD257 pKa = 3.54ASIIAAQVDD266 pKa = 3.79NGSNDD271 pKa = 3.12ACGIASMTVVPNTFDD286 pKa = 3.67CSNIGANTVTLTVVDD301 pKa = 3.97NNGNSSSATATVTVNDD317 pKa = 3.42VTVAQVVTQDD327 pKa = 3.2ISIDD331 pKa = 3.54LDD333 pKa = 3.6ASGDD337 pKa = 3.58ASIIAAQVDD346 pKa = 4.1DD347 pKa = 4.94GSNDD351 pKa = 3.14ACGIASMTVVPNTFDD366 pKa = 3.67CSNIGANTVTLTVVDD381 pKa = 4.02VNGNSSSATATVTVNDD397 pKa = 3.3VTAAQVITQDD407 pKa = 3.52ISIDD411 pKa = 3.36LDD413 pKa = 3.95ANGDD417 pKa = 3.54ASIIAAQVDD426 pKa = 3.79NGSNDD431 pKa = 3.12ACGIASMTVVPNTFDD446 pKa = 3.67CSNIGANTVTLTVVDD461 pKa = 3.97NNGNSSSATATVTVNDD477 pKa = 3.3VTAAQVITQNTSIDD491 pKa = 3.4LDD493 pKa = 4.08ANGNASIVPADD504 pKa = 3.8VDD506 pKa = 3.65NGSNDD511 pKa = 3.17ACGIASMTVTPNTFDD526 pKa = 3.9CSNIGANTVTLTVVDD541 pKa = 4.76LNGNSNSAMATVTVNDD557 pKa = 3.93VIAAQVITQNISLDD571 pKa = 3.42LDD573 pKa = 4.15ANGGASIVPADD584 pKa = 3.9VDD586 pKa = 3.79NGSSDD591 pKa = 3.37ACGIASMTVTPNTFDD606 pKa = 3.7CSNVGANTVTLTVTDD621 pKa = 3.47VNGNVSSATANVTVNDD637 pKa = 3.63ITPAIVITQNTSIDD651 pKa = 3.4LDD653 pKa = 4.08ANGNASIVPADD664 pKa = 3.8VDD666 pKa = 3.65NGSNDD671 pKa = 3.04ACGIANMTVSPNTFDD686 pKa = 4.69CSNLGMNTVTLTVTDD701 pKa = 3.66VNGNISSATAVVSISDD717 pKa = 3.48NTAPDD722 pKa = 3.93IVCVPDD728 pKa = 3.5GTRR731 pKa = 11.84DD732 pKa = 3.56TDD734 pKa = 3.91PGQCTYY740 pKa = 10.04TIQGAEE746 pKa = 3.89FDD748 pKa = 5.84AIFTDD753 pKa = 3.86NCSSGVITNSINGTATLAGEE773 pKa = 4.58VFMQGATEE781 pKa = 4.21VTWIVDD787 pKa = 3.59DD788 pKa = 4.21GHH790 pKa = 6.55GQTASCTTTITVEE803 pKa = 4.14DD804 pKa = 4.04NEE806 pKa = 4.43APIVDD811 pKa = 5.13CINIQVLLDD820 pKa = 3.46ASGNGTITVADD831 pKa = 3.82INNNSTDD838 pKa = 3.13NCGIASITLSQTSFDD853 pKa = 4.26CSDD856 pKa = 2.98IGGDD860 pKa = 3.74LDD862 pKa = 3.71EE863 pKa = 6.1LIISEE868 pKa = 4.29YY869 pKa = 11.04SDD871 pKa = 3.13GTGNNDD877 pKa = 4.48CIEE880 pKa = 4.37IYY882 pKa = 10.71NGTGNPINLYY892 pKa = 10.8AEE894 pKa = 5.54GYY896 pKa = 6.78TLRR899 pKa = 11.84FYY901 pKa = 11.72YY902 pKa = 10.49NGSTSYY908 pKa = 9.54TQVPLLGTVADD919 pKa = 3.98RR920 pKa = 11.84EE921 pKa = 4.72VYY923 pKa = 9.54VVCNPFGPGAMQADD937 pKa = 3.7QTGNFGFDD945 pKa = 3.27GNDD948 pKa = 3.47AIALTKK954 pKa = 10.45AGNAIDD960 pKa = 4.31VIGQIGVNPGTGWTVGSNSTAGTTLVRR987 pKa = 11.84NKK989 pKa = 10.56DD990 pKa = 3.51VLYY993 pKa = 11.3GNINDD998 pKa = 3.49ISGISSEE1005 pKa = 3.94WTQYY1009 pKa = 9.75AQNNTSNLGSHH1020 pKa = 6.87EE1021 pKa = 4.1IEE1023 pKa = 4.38IADD1026 pKa = 3.86LAKK1029 pKa = 10.79NVILTVTDD1037 pKa = 3.2TSGNVSTCEE1046 pKa = 3.96GNVTVIDD1053 pKa = 4.45NIAPVAMCQSVTIQLDD1069 pKa = 3.72ANGVASVTASEE1080 pKa = 4.45VDD1082 pKa = 3.22NGSNDD1087 pKa = 3.16ACGIASLVLDD1097 pKa = 3.75QTDD1100 pKa = 4.32FTCANLGDD1108 pKa = 3.86NTVILTVTDD1117 pKa = 3.61NNGNSSSCEE1126 pKa = 3.62ATITVEE1132 pKa = 4.75DD1133 pKa = 4.69NIDD1136 pKa = 3.82PTVMTQDD1143 pKa = 3.64LTIQLDD1149 pKa = 4.07ANGSASITPADD1160 pKa = 4.14IDD1162 pKa = 3.69NGSIDD1167 pKa = 3.49NCGIATQTVTPNSFDD1182 pKa = 3.59CSNVGVNTVTLTVTDD1197 pKa = 3.44TSGNVSTGIATVTVEE1212 pKa = 5.14DD1213 pKa = 3.93NVAPIAICKK1222 pKa = 10.16NITVEE1227 pKa = 4.28LGINGEE1233 pKa = 4.13ATITGADD1240 pKa = 3.56VDD1242 pKa = 4.41NGSSDD1247 pKa = 3.35ACGIASLDD1255 pKa = 3.4VSPSVFGCADD1265 pKa = 3.02IGANTVTLTVTDD1277 pKa = 3.61NNGNVSTCSATVTVTGIVPVVTISQGPLPEE1307 pKa = 4.45FCQGAVLVLTAEE1319 pKa = 4.09SDD1321 pKa = 3.44EE1322 pKa = 4.41DD1323 pKa = 5.2FGYY1326 pKa = 11.06LWTTGEE1332 pKa = 4.34TTQSIEE1338 pKa = 3.71IPGNGTYY1345 pKa = 10.78GVTVTSLTNCSTEE1358 pKa = 3.97AEE1360 pKa = 4.08ITITGFNAGVLVSSYY1375 pKa = 10.74TIIASEE1381 pKa = 4.28KK1382 pKa = 10.43VEE1384 pKa = 4.19LKK1386 pKa = 11.13NNVIVQTGGVGVTKK1400 pKa = 10.68ASGEE1404 pKa = 4.08IKK1406 pKa = 9.92IEE1408 pKa = 4.05KK1409 pKa = 9.54ASHH1412 pKa = 5.5IVGLGQANKK1421 pKa = 10.13IEE1423 pKa = 4.28IKK1425 pKa = 10.28QGSSVGTAVYY1435 pKa = 10.33QPANPTIPAFVYY1447 pKa = 10.26NGYY1450 pKa = 10.08SSSSSPDD1457 pKa = 2.81VRR1459 pKa = 11.84VNNNQTLTLSGGVYY1473 pKa = 10.46NKK1475 pKa = 10.06VEE1477 pKa = 4.23LEE1479 pKa = 4.08EE1480 pKa = 4.19NATVLFTASNVYY1492 pKa = 10.3INEE1495 pKa = 4.17LKK1497 pKa = 9.52TKK1499 pKa = 10.22KK1500 pKa = 10.33SATIEE1505 pKa = 4.12FNGCTNVFLNKK1516 pKa = 9.67EE1517 pKa = 4.22FKK1519 pKa = 10.89LDD1521 pKa = 3.79DD1522 pKa = 3.97NGVFNSNGYY1531 pKa = 8.33MVTVYY1536 pKa = 10.59INDD1539 pKa = 3.52DD1540 pKa = 3.87FEE1542 pKa = 4.69VKK1544 pKa = 10.25KK1545 pKa = 11.2GSDD1548 pKa = 3.23FTGRR1552 pKa = 11.84VHH1554 pKa = 6.79TNNHH1558 pKa = 6.24DD1559 pKa = 3.7IEE1561 pKa = 4.83VEE1563 pKa = 4.11GHH1565 pKa = 5.97NSSTTYY1571 pKa = 7.51MTGLFIGKK1579 pKa = 9.26KK1580 pKa = 9.73VEE1582 pKa = 3.81ADD1584 pKa = 3.84KK1585 pKa = 11.54NVVWNADD1592 pKa = 3.95QYY1594 pKa = 11.69CDD1596 pKa = 3.93PCPIEE1601 pKa = 5.2APINNGGSEE1610 pKa = 4.8PITDD1614 pKa = 3.53YY1615 pKa = 11.25TIIAFDD1621 pKa = 3.88EE1622 pKa = 4.46VHH1624 pKa = 6.27LHH1626 pKa = 6.69GDD1628 pKa = 3.73NIVQTGGVGVTRR1640 pKa = 11.84HH1641 pKa = 5.19KK1642 pKa = 11.0KK1643 pKa = 9.89KK1644 pKa = 10.53IKK1646 pKa = 9.35LHH1648 pKa = 6.22KK1649 pKa = 9.85DD1650 pKa = 2.95SHH1652 pKa = 5.22ITEE1655 pKa = 4.38FAKK1658 pKa = 10.7ASQIQVNGGSSIGTRR1673 pKa = 11.84ILSPADD1679 pKa = 3.82PIIPLFVKK1687 pKa = 10.38NVYY1690 pKa = 10.51SNSSSPNATINSGQTVTLTGSVYY1713 pKa = 11.08DD1714 pKa = 4.39KK1715 pKa = 11.58VDD1717 pKa = 3.72LKK1719 pKa = 11.24EE1720 pKa = 3.97GATVIFAQSNVFINEE1735 pKa = 3.91LKK1737 pKa = 7.7TRR1739 pKa = 11.84KK1740 pKa = 9.39YY1741 pKa = 10.22ATIKK1745 pKa = 10.71FSGCTNVFIKK1755 pKa = 10.77KK1756 pKa = 8.76EE1757 pKa = 4.14FKK1759 pKa = 10.61LDD1761 pKa = 3.62DD1762 pKa = 3.85YY1763 pKa = 11.89GVINPEE1769 pKa = 3.86TDD1771 pKa = 2.59GHH1773 pKa = 5.84KK1774 pKa = 10.7VVFHH1778 pKa = 6.73VDD1780 pKa = 3.25DD1781 pKa = 4.75DD1782 pKa = 4.58VEE1784 pKa = 4.12IDD1786 pKa = 3.31KK1787 pKa = 11.39GSVVVASIYY1796 pKa = 10.69AYY1798 pKa = 10.64NDD1800 pKa = 3.41EE1801 pKa = 4.41IKK1803 pKa = 10.76VDD1805 pKa = 4.09GSSSDD1810 pKa = 3.68PVSMKK1815 pKa = 10.87GLFIAKK1821 pKa = 9.21KK1822 pKa = 8.11VHH1824 pKa = 5.15AHH1826 pKa = 7.15EE1827 pKa = 4.85DD1828 pKa = 3.75VLWNKK1833 pKa = 8.19DD1834 pKa = 3.45TSGSPCPVITPTAQAPVAPEE1854 pKa = 3.6VGNRR1858 pKa = 11.84SIEE1861 pKa = 4.39LISVKK1866 pKa = 10.18AWPNPSDD1873 pKa = 3.44TVFNLRR1879 pKa = 11.84LITEE1883 pKa = 4.13NQVDD1887 pKa = 4.25VVSILVFDD1895 pKa = 4.49SNNKK1899 pKa = 9.0LVHH1902 pKa = 6.35SSTFRR1907 pKa = 11.84PEE1909 pKa = 3.46AVYY1912 pKa = 10.61QFGNEE1917 pKa = 4.11LEE1919 pKa = 4.24SGVYY1923 pKa = 8.69IVKK1926 pKa = 10.19VSQAGSNAYY1935 pKa = 8.71TRR1937 pKa = 11.84VIKK1940 pKa = 10.41FF1941 pKa = 3.24
Molecular weight: 200.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q0XBG4|A0A4Q0XBG4_9FLAO Queuine tRNA-ribosyltransferase OS=Gelidibacter gilvus OX=59602 GN=tgt PE=3 SV=1
MM1 pKa = 7.05TRR3 pKa = 11.84GIRR6 pKa = 11.84FSVGSLQFAVFSWQSSVGSLQLAVFSWQSSVGSLQLAVFRR46 pKa = 11.84RR47 pKa = 11.84LL48 pKa = 3.16
MM1 pKa = 7.05TRR3 pKa = 11.84GIRR6 pKa = 11.84FSVGSLQFAVFSWQSSVGSLQLAVFSWQSSVGSLQLAVFRR46 pKa = 11.84RR47 pKa = 11.84LL48 pKa = 3.16
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1309719 |
24 |
3758 |
350.9 |
39.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.446 ± 0.037 | 0.695 ± 0.013 |
5.821 ± 0.029 | 6.442 ± 0.035 |
5.204 ± 0.028 | 6.328 ± 0.037 |
1.951 ± 0.019 | 7.91 ± 0.037 |
7.529 ± 0.043 | 9.329 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.345 ± 0.021 | 5.948 ± 0.042 |
3.447 ± 0.022 | 3.447 ± 0.019 |
3.523 ± 0.024 | 6.552 ± 0.034 |
5.794 ± 0.044 | 6.181 ± 0.03 |
1.052 ± 0.013 | 4.057 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
