
Salmon aquaparamyxovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Aquaparamyxovirus
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
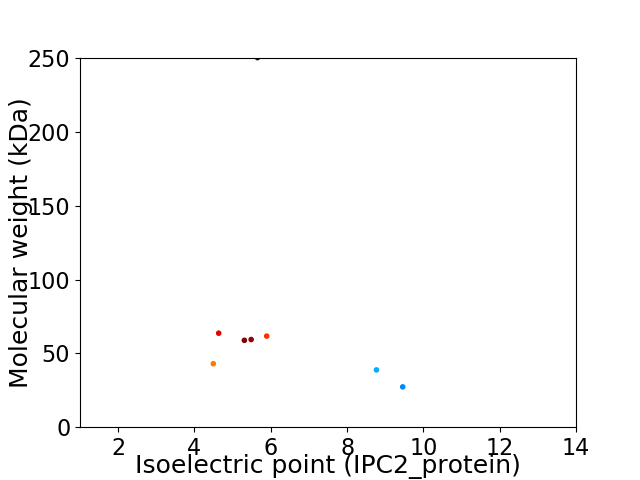
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G2KTE9|A0A3G2KTE9_9MONO Hemagglutinin-neuraminidase protein OS=Salmon aquaparamyxovirus OX=381543 GN=HN PE=3 SV=1
MM1 pKa = 7.4SSYY4 pKa = 10.73GRR6 pKa = 11.84SAGKK10 pKa = 10.04DD11 pKa = 3.15KK12 pKa = 10.89QVEE15 pKa = 4.08RR16 pKa = 11.84QEE18 pKa = 4.41TSRR21 pKa = 11.84HH22 pKa = 4.81PNEE25 pKa = 4.52RR26 pKa = 11.84EE27 pKa = 3.87SLLDD31 pKa = 3.56SGLSNIQSLLRR42 pKa = 11.84DD43 pKa = 3.75INSSTRR49 pKa = 11.84TSQKK53 pKa = 10.68KK54 pKa = 9.84KK55 pKa = 10.25LNSGADD61 pKa = 3.92DD62 pKa = 3.63TSSTHH67 pKa = 6.06SASSSDD73 pKa = 3.53RR74 pKa = 11.84PRR76 pKa = 11.84DD77 pKa = 3.72NAGGTSDD84 pKa = 3.65RR85 pKa = 11.84RR86 pKa = 11.84VRR88 pKa = 11.84SQSQDD93 pKa = 2.47AVTQRR98 pKa = 11.84DD99 pKa = 3.86ISSLHH104 pKa = 6.58CDD106 pKa = 3.63DD107 pKa = 5.06PRR109 pKa = 11.84SQVNCSANKK118 pKa = 10.16RR119 pKa = 11.84STGPPAPQPRR129 pKa = 11.84GRR131 pKa = 11.84DD132 pKa = 3.29LHH134 pKa = 7.54KK135 pKa = 10.94GVVGVNEE142 pKa = 3.87IPPYY146 pKa = 10.47EE147 pKa = 5.12GGDD150 pKa = 3.12VDD152 pKa = 3.84QLVNEE157 pKa = 4.54EE158 pKa = 4.31EE159 pKa = 4.25EE160 pKa = 4.45ADD162 pKa = 3.39IPMAEE167 pKa = 4.21QQDD170 pKa = 4.29GADD173 pKa = 3.6EE174 pKa = 4.11EE175 pKa = 5.38SYY177 pKa = 10.92PEE179 pKa = 3.75QAGRR183 pKa = 11.84GSSARR188 pKa = 11.84SSLGGGSALAGWGRR202 pKa = 11.84QANTSSPAEE211 pKa = 3.8ISEE214 pKa = 4.71GYY216 pKa = 10.8DD217 pKa = 4.33DD218 pKa = 3.89VEE220 pKa = 4.08RR221 pKa = 11.84WAKK224 pKa = 10.3HH225 pKa = 4.13ATLPEE230 pKa = 4.38LFTRR234 pKa = 11.84VQSGPNGVCSPVAIVAEE251 pKa = 4.45EE252 pKa = 3.96AQLLYY257 pKa = 10.14TVEE260 pKa = 4.51PEE262 pKa = 3.84PVMSEE267 pKa = 4.02EE268 pKa = 4.15EE269 pKa = 3.77EE270 pKa = 4.41AMIDD274 pKa = 3.63QALNDD279 pKa = 4.22TLDD282 pKa = 3.36VDD284 pKa = 4.75SDD286 pKa = 3.97GGDD289 pKa = 3.49DD290 pKa = 4.25SPLIEE295 pKa = 4.3QMSVLLDD302 pKa = 5.88DD303 pKa = 4.07EE304 pKa = 5.17QEE306 pKa = 4.22WNNLDD311 pKa = 4.12QNRR314 pKa = 11.84GDD316 pKa = 3.34QDD318 pKa = 3.87QNQEE322 pKa = 4.11WKK324 pKa = 10.72DD325 pKa = 3.42SALNIGKK332 pKa = 9.62KK333 pKa = 6.65GHH335 pKa = 5.85RR336 pKa = 11.84RR337 pKa = 11.84EE338 pKa = 3.8VSIYY342 pKa = 8.27TRR344 pKa = 11.84NGICYY349 pKa = 9.97LDD351 pKa = 3.68TWCNPCCVPIRR362 pKa = 11.84VHH364 pKa = 5.37PTRR367 pKa = 11.84EE368 pKa = 3.5RR369 pKa = 11.84CICRR373 pKa = 11.84LCPEE377 pKa = 4.31TCGDD381 pKa = 3.86CAVATEE387 pKa = 4.95DD388 pKa = 3.73PTIPSII394 pKa = 4.6
MM1 pKa = 7.4SSYY4 pKa = 10.73GRR6 pKa = 11.84SAGKK10 pKa = 10.04DD11 pKa = 3.15KK12 pKa = 10.89QVEE15 pKa = 4.08RR16 pKa = 11.84QEE18 pKa = 4.41TSRR21 pKa = 11.84HH22 pKa = 4.81PNEE25 pKa = 4.52RR26 pKa = 11.84EE27 pKa = 3.87SLLDD31 pKa = 3.56SGLSNIQSLLRR42 pKa = 11.84DD43 pKa = 3.75INSSTRR49 pKa = 11.84TSQKK53 pKa = 10.68KK54 pKa = 9.84KK55 pKa = 10.25LNSGADD61 pKa = 3.92DD62 pKa = 3.63TSSTHH67 pKa = 6.06SASSSDD73 pKa = 3.53RR74 pKa = 11.84PRR76 pKa = 11.84DD77 pKa = 3.72NAGGTSDD84 pKa = 3.65RR85 pKa = 11.84RR86 pKa = 11.84VRR88 pKa = 11.84SQSQDD93 pKa = 2.47AVTQRR98 pKa = 11.84DD99 pKa = 3.86ISSLHH104 pKa = 6.58CDD106 pKa = 3.63DD107 pKa = 5.06PRR109 pKa = 11.84SQVNCSANKK118 pKa = 10.16RR119 pKa = 11.84STGPPAPQPRR129 pKa = 11.84GRR131 pKa = 11.84DD132 pKa = 3.29LHH134 pKa = 7.54KK135 pKa = 10.94GVVGVNEE142 pKa = 3.87IPPYY146 pKa = 10.47EE147 pKa = 5.12GGDD150 pKa = 3.12VDD152 pKa = 3.84QLVNEE157 pKa = 4.54EE158 pKa = 4.31EE159 pKa = 4.25EE160 pKa = 4.45ADD162 pKa = 3.39IPMAEE167 pKa = 4.21QQDD170 pKa = 4.29GADD173 pKa = 3.6EE174 pKa = 4.11EE175 pKa = 5.38SYY177 pKa = 10.92PEE179 pKa = 3.75QAGRR183 pKa = 11.84GSSARR188 pKa = 11.84SSLGGGSALAGWGRR202 pKa = 11.84QANTSSPAEE211 pKa = 3.8ISEE214 pKa = 4.71GYY216 pKa = 10.8DD217 pKa = 4.33DD218 pKa = 3.89VEE220 pKa = 4.08RR221 pKa = 11.84WAKK224 pKa = 10.3HH225 pKa = 4.13ATLPEE230 pKa = 4.38LFTRR234 pKa = 11.84VQSGPNGVCSPVAIVAEE251 pKa = 4.45EE252 pKa = 3.96AQLLYY257 pKa = 10.14TVEE260 pKa = 4.51PEE262 pKa = 3.84PVMSEE267 pKa = 4.02EE268 pKa = 4.15EE269 pKa = 3.77EE270 pKa = 4.41AMIDD274 pKa = 3.63QALNDD279 pKa = 4.22TLDD282 pKa = 3.36VDD284 pKa = 4.75SDD286 pKa = 3.97GGDD289 pKa = 3.49DD290 pKa = 4.25SPLIEE295 pKa = 4.3QMSVLLDD302 pKa = 5.88DD303 pKa = 4.07EE304 pKa = 5.17QEE306 pKa = 4.22WNNLDD311 pKa = 4.12QNRR314 pKa = 11.84GDD316 pKa = 3.34QDD318 pKa = 3.87QNQEE322 pKa = 4.11WKK324 pKa = 10.72DD325 pKa = 3.42SALNIGKK332 pKa = 9.62KK333 pKa = 6.65GHH335 pKa = 5.85RR336 pKa = 11.84RR337 pKa = 11.84EE338 pKa = 3.8VSIYY342 pKa = 8.27TRR344 pKa = 11.84NGICYY349 pKa = 9.97LDD351 pKa = 3.68TWCNPCCVPIRR362 pKa = 11.84VHH364 pKa = 5.37PTRR367 pKa = 11.84EE368 pKa = 3.5RR369 pKa = 11.84CICRR373 pKa = 11.84LCPEE377 pKa = 4.31TCGDD381 pKa = 3.86CAVATEE387 pKa = 4.95DD388 pKa = 3.73PTIPSII394 pKa = 4.6
Molecular weight: 43.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2KTD2|A0A3G2KTD2_9MONO V protein OS=Salmon aquaparamyxovirus OX=381543 GN=V PE=4 SV=1
MM1 pKa = 7.72AGLLARR7 pKa = 11.84INKK10 pKa = 8.09WRR12 pKa = 11.84GKK14 pKa = 9.55KK15 pKa = 9.52QADD18 pKa = 3.44TPMRR22 pKa = 11.84EE23 pKa = 4.31SLYY26 pKa = 9.78STQDD30 pKa = 3.02FPTFSPSSEE39 pKa = 3.7ISTPPPEE46 pKa = 4.27PPKK49 pKa = 10.5RR50 pKa = 11.84RR51 pKa = 11.84NLTVVPMTPRR61 pKa = 11.84APTPPQVLTGPGTMLVVHH79 pKa = 7.09PIGGYY84 pKa = 8.88DD85 pKa = 3.61PKK87 pKa = 11.27AKK89 pKa = 9.35TLSPSGILAAYY100 pKa = 8.99IATIQGARR108 pKa = 11.84SIAVLTRR115 pKa = 11.84DD116 pKa = 4.06LQDD119 pKa = 3.68LLLRR123 pKa = 11.84SPEE126 pKa = 3.63EE127 pKa = 4.21GIFIRR132 pKa = 11.84EE133 pKa = 4.09LLEE136 pKa = 3.8STRR139 pKa = 11.84YY140 pKa = 10.28LLMRR144 pKa = 11.84VEE146 pKa = 4.32MWISLLTRR154 pKa = 11.84KK155 pKa = 9.35KK156 pKa = 10.43RR157 pKa = 11.84PTYY160 pKa = 9.16QWLNNRR166 pKa = 11.84MEE168 pKa = 4.04QMKK171 pKa = 10.25RR172 pKa = 11.84VTLNKK177 pKa = 9.82PVEE180 pKa = 4.38DD181 pKa = 4.53LLLDD185 pKa = 3.75QAWVVVVPWLDD196 pKa = 3.41GEE198 pKa = 4.72GRR200 pKa = 11.84QILPARR206 pKa = 11.84QKK208 pKa = 10.57SLRR211 pKa = 11.84DD212 pKa = 3.3MTTLRR217 pKa = 11.84GGQSMQHH224 pKa = 5.43SQSFSPASRR233 pKa = 11.84ADD235 pKa = 3.42RR236 pKa = 11.84TEE238 pKa = 3.84YY239 pKa = 10.28VAQQ242 pKa = 3.89
MM1 pKa = 7.72AGLLARR7 pKa = 11.84INKK10 pKa = 8.09WRR12 pKa = 11.84GKK14 pKa = 9.55KK15 pKa = 9.52QADD18 pKa = 3.44TPMRR22 pKa = 11.84EE23 pKa = 4.31SLYY26 pKa = 9.78STQDD30 pKa = 3.02FPTFSPSSEE39 pKa = 3.7ISTPPPEE46 pKa = 4.27PPKK49 pKa = 10.5RR50 pKa = 11.84RR51 pKa = 11.84NLTVVPMTPRR61 pKa = 11.84APTPPQVLTGPGTMLVVHH79 pKa = 7.09PIGGYY84 pKa = 8.88DD85 pKa = 3.61PKK87 pKa = 11.27AKK89 pKa = 9.35TLSPSGILAAYY100 pKa = 8.99IATIQGARR108 pKa = 11.84SIAVLTRR115 pKa = 11.84DD116 pKa = 4.06LQDD119 pKa = 3.68LLLRR123 pKa = 11.84SPEE126 pKa = 3.63EE127 pKa = 4.21GIFIRR132 pKa = 11.84EE133 pKa = 4.09LLEE136 pKa = 3.8STRR139 pKa = 11.84YY140 pKa = 10.28LLMRR144 pKa = 11.84VEE146 pKa = 4.32MWISLLTRR154 pKa = 11.84KK155 pKa = 9.35KK156 pKa = 10.43RR157 pKa = 11.84PTYY160 pKa = 9.16QWLNNRR166 pKa = 11.84MEE168 pKa = 4.04QMKK171 pKa = 10.25RR172 pKa = 11.84VTLNKK177 pKa = 9.82PVEE180 pKa = 4.38DD181 pKa = 4.53LLLDD185 pKa = 3.75QAWVVVVPWLDD196 pKa = 3.41GEE198 pKa = 4.72GRR200 pKa = 11.84QILPARR206 pKa = 11.84QKK208 pKa = 10.57SLRR211 pKa = 11.84DD212 pKa = 3.3MTTLRR217 pKa = 11.84GGQSMQHH224 pKa = 5.43SQSFSPASRR233 pKa = 11.84ADD235 pKa = 3.42RR236 pKa = 11.84TEE238 pKa = 3.84YY239 pKa = 10.28VAQQ242 pKa = 3.89
Molecular weight: 27.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5450 |
242 |
2213 |
681.3 |
75.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.294 ± 0.385 | 1.945 ± 0.273 |
6.11 ± 0.43 | 5.89 ± 0.574 |
2.917 ± 0.655 | 7.009 ± 0.615 |
1.725 ± 0.231 | 6.018 ± 0.586 |
5.138 ± 0.487 | 9.156 ± 0.463 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.789 ± 0.427 | 4.422 ± 0.336 |
4.899 ± 0.854 | 3.927 ± 0.369 |
5.266 ± 0.506 | 8.661 ± 0.786 |
6.312 ± 0.29 | 7.064 ± 0.362 |
1.229 ± 0.169 | 3.229 ± 0.343 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
