
Aureimonas jatrophae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Aureimonas
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
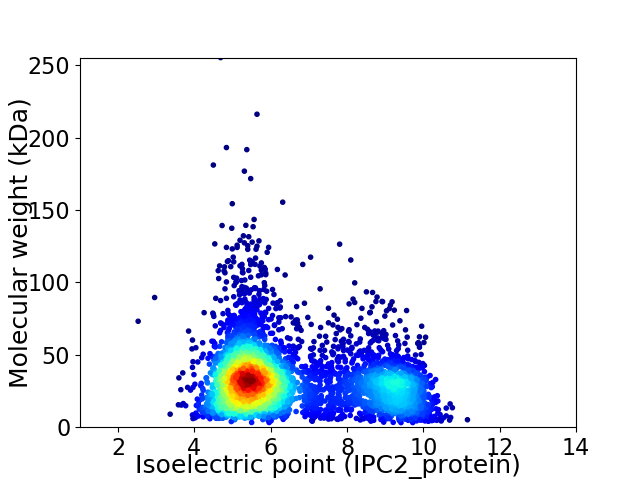
Virtual 2D-PAGE plot for 4295 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
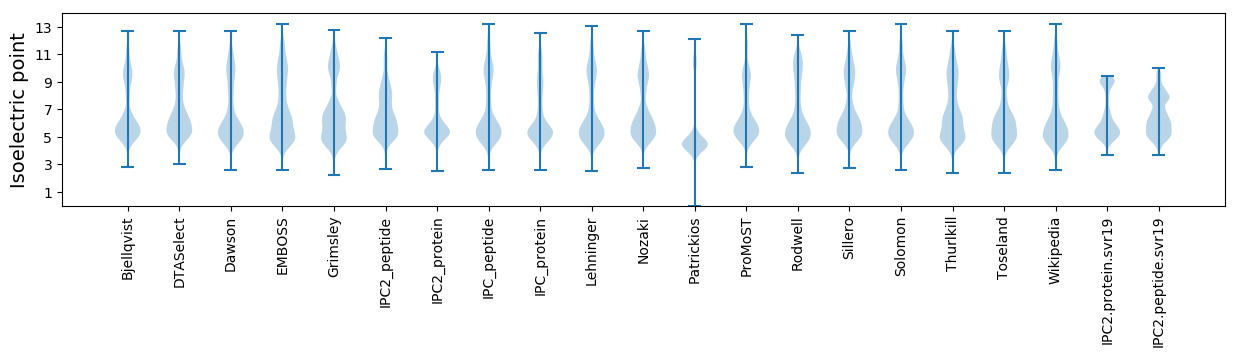
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0K3Y5|A0A1H0K3Y5_9RHIZ Voltage-gated potassium channel OS=Aureimonas jatrophae OX=1166073 GN=SAMN05192530_10779 PE=4 SV=1
MM1 pKa = 7.32AAAYY5 pKa = 7.96TGTRR9 pKa = 11.84WGASDD14 pKa = 4.13AAGTPGGVVTWSFNLLGAEE33 pKa = 3.92FFGFDD38 pKa = 4.08EE39 pKa = 5.66SILSAPFQAAVRR51 pKa = 11.84AAFDD55 pKa = 3.33VWEE58 pKa = 4.31SLANIDD64 pKa = 3.89FQEE67 pKa = 4.19VATASAQIQLGWDD80 pKa = 3.49TFDD83 pKa = 3.84GAGGTLALAYY93 pKa = 8.44WQYY96 pKa = 11.61QGAKK100 pKa = 8.06TLQAEE105 pKa = 4.49VAFDD109 pKa = 4.06IAEE112 pKa = 4.12NWNPTAGGASFQAVAIHH129 pKa = 6.67EE130 pKa = 4.43IGHH133 pKa = 6.83AIGLAHH139 pKa = 6.92TDD141 pKa = 3.65DD142 pKa = 4.77PTSIMYY148 pKa = 8.75PYY150 pKa = 10.71LSAQTLPSASDD161 pKa = 2.95IAAIQGLYY169 pKa = 10.24GPSDD173 pKa = 3.42RR174 pKa = 11.84GFSLPGTAGNDD185 pKa = 2.84ILTGGRR191 pKa = 11.84GNDD194 pKa = 3.5VISGLEE200 pKa = 4.07GADD203 pKa = 3.49TLQGGLGRR211 pKa = 11.84DD212 pKa = 3.5VLQGNQGDD220 pKa = 4.43DD221 pKa = 3.75LVQGGHH227 pKa = 7.09DD228 pKa = 3.7GDD230 pKa = 4.3EE231 pKa = 4.32VSGGQGNDD239 pKa = 2.63WVYY242 pKa = 11.67GNLGNDD248 pKa = 3.56TLSGNLGNDD257 pKa = 3.42VLSGGAGADD266 pKa = 3.07TFVFMPGSGADD277 pKa = 2.9IVTDD281 pKa = 3.55YY282 pKa = 11.75AFAEE286 pKa = 4.35GDD288 pKa = 3.53RR289 pKa = 11.84LNVGGRR295 pKa = 11.84TYY297 pKa = 11.42AVATASDD304 pKa = 4.33GSALLQLADD313 pKa = 3.62GDD315 pKa = 4.41IIILQGVTQGEE326 pKa = 4.58FSQSYY331 pKa = 7.79VAA333 pKa = 5.39
MM1 pKa = 7.32AAAYY5 pKa = 7.96TGTRR9 pKa = 11.84WGASDD14 pKa = 4.13AAGTPGGVVTWSFNLLGAEE33 pKa = 3.92FFGFDD38 pKa = 4.08EE39 pKa = 5.66SILSAPFQAAVRR51 pKa = 11.84AAFDD55 pKa = 3.33VWEE58 pKa = 4.31SLANIDD64 pKa = 3.89FQEE67 pKa = 4.19VATASAQIQLGWDD80 pKa = 3.49TFDD83 pKa = 3.84GAGGTLALAYY93 pKa = 8.44WQYY96 pKa = 11.61QGAKK100 pKa = 8.06TLQAEE105 pKa = 4.49VAFDD109 pKa = 4.06IAEE112 pKa = 4.12NWNPTAGGASFQAVAIHH129 pKa = 6.67EE130 pKa = 4.43IGHH133 pKa = 6.83AIGLAHH139 pKa = 6.92TDD141 pKa = 3.65DD142 pKa = 4.77PTSIMYY148 pKa = 8.75PYY150 pKa = 10.71LSAQTLPSASDD161 pKa = 2.95IAAIQGLYY169 pKa = 10.24GPSDD173 pKa = 3.42RR174 pKa = 11.84GFSLPGTAGNDD185 pKa = 2.84ILTGGRR191 pKa = 11.84GNDD194 pKa = 3.5VISGLEE200 pKa = 4.07GADD203 pKa = 3.49TLQGGLGRR211 pKa = 11.84DD212 pKa = 3.5VLQGNQGDD220 pKa = 4.43DD221 pKa = 3.75LVQGGHH227 pKa = 7.09DD228 pKa = 3.7GDD230 pKa = 4.3EE231 pKa = 4.32VSGGQGNDD239 pKa = 2.63WVYY242 pKa = 11.67GNLGNDD248 pKa = 3.56TLSGNLGNDD257 pKa = 3.42VLSGGAGADD266 pKa = 3.07TFVFMPGSGADD277 pKa = 2.9IVTDD281 pKa = 3.55YY282 pKa = 11.75AFAEE286 pKa = 4.35GDD288 pKa = 3.53RR289 pKa = 11.84LNVGGRR295 pKa = 11.84TYY297 pKa = 11.42AVATASDD304 pKa = 4.33GSALLQLADD313 pKa = 3.62GDD315 pKa = 4.41IIILQGVTQGEE326 pKa = 4.58FSQSYY331 pKa = 7.79VAA333 pKa = 5.39
Molecular weight: 34.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0KQ96|A0A1H0KQ96_9RHIZ Uncharacterized protein OS=Aureimonas jatrophae OX=1166073 GN=SAMN05192530_10890 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1370096 |
31 |
2376 |
319.0 |
34.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.222 ± 0.046 | 0.735 ± 0.011 |
5.798 ± 0.03 | 5.891 ± 0.036 |
3.651 ± 0.025 | 8.902 ± 0.034 |
1.901 ± 0.021 | 4.531 ± 0.024 |
2.231 ± 0.028 | 10.472 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.23 ± 0.015 | 2.231 ± 0.021 |
5.39 ± 0.029 | 2.835 ± 0.021 |
8.377 ± 0.042 | 5.303 ± 0.03 |
5.307 ± 0.028 | 7.795 ± 0.03 |
1.246 ± 0.015 | 1.952 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
