
Parietaria mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
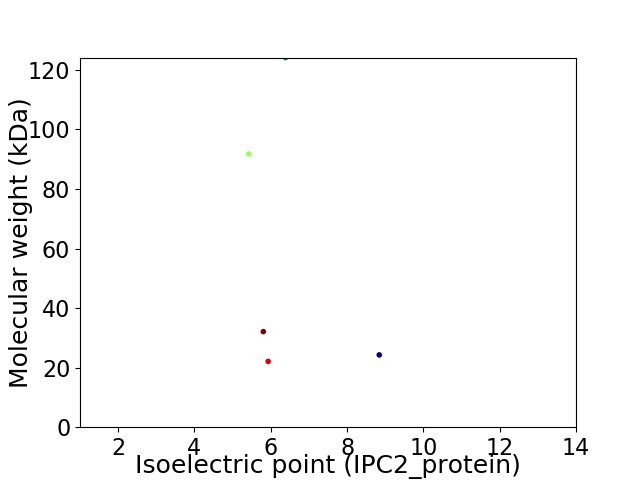
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
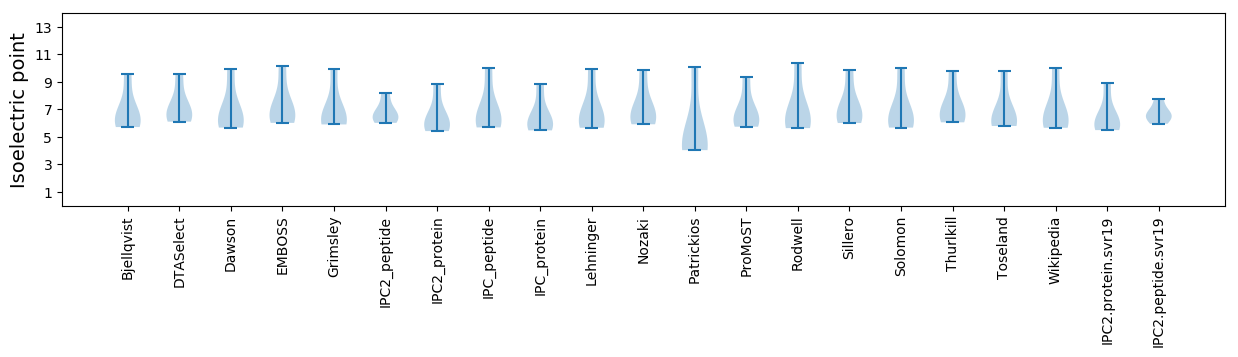
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6RK14|Q6RK14_9BROM ATP-dependent helicase OS=Parietaria mottle virus OX=64958 PE=3 SV=1
MM1 pKa = 7.42DD2 pKa = 4.85QIRR5 pKa = 11.84INLIDD10 pKa = 3.85SFLSSRR16 pKa = 11.84LCFNATFGVYY26 pKa = 9.97HH27 pKa = 7.73DD28 pKa = 6.31DD29 pKa = 3.44YY30 pKa = 10.91MEE32 pKa = 4.05WVKK35 pKa = 10.95LFLFKK40 pKa = 10.7HH41 pKa = 5.64VIEE44 pKa = 4.61HH45 pKa = 5.19TAKK48 pKa = 10.22FDD50 pKa = 3.39FATIHH55 pKa = 5.5MTLEE59 pKa = 4.03MVLCRR64 pKa = 11.84LVPGLGDD71 pKa = 3.31ALYY74 pKa = 9.94PEE76 pKa = 5.17EE77 pKa = 4.19EE78 pKa = 4.37NEE80 pKa = 4.03VDD82 pKa = 3.52GLSSRR87 pKa = 11.84EE88 pKa = 3.48IDD90 pKa = 3.94PFYY93 pKa = 11.13LPYY96 pKa = 10.75DD97 pKa = 4.42DD98 pKa = 6.49LDD100 pKa = 3.52VDD102 pKa = 3.87YY103 pKa = 9.26TALCRR108 pKa = 11.84EE109 pKa = 4.42RR110 pKa = 11.84DD111 pKa = 3.71DD112 pKa = 5.27SIPTIPPEE120 pKa = 4.47DD121 pKa = 4.05EE122 pKa = 4.25LSVIVKK128 pKa = 10.05NMTYY132 pKa = 9.91IPEE135 pKa = 4.38GTSWGDD141 pKa = 3.23VSDD144 pKa = 5.25SSFQEE149 pKa = 4.01HH150 pKa = 6.79LEE152 pKa = 4.34EE153 pKa = 4.35IQPVPLGSDD162 pKa = 3.25VGHH165 pKa = 7.18TIPNEE170 pKa = 4.31HH171 pKa = 7.05IPFDD175 pKa = 5.07DD176 pKa = 5.19DD177 pKa = 3.8GQLVDD182 pKa = 4.54VQWSDD187 pKa = 4.77AIPSLATDD195 pKa = 3.85VTCDD199 pKa = 3.04ADD201 pKa = 3.62YY202 pKa = 10.99RR203 pKa = 11.84EE204 pKa = 4.92CGFVSYY210 pKa = 11.05SSGSQHH216 pKa = 6.44SSRR219 pKa = 11.84WKK221 pKa = 9.83PKK223 pKa = 9.11VSQVKK228 pKa = 9.2PDD230 pKa = 3.64PSIIQSAIDD239 pKa = 3.46EE240 pKa = 5.0LFPHH244 pKa = 6.27HH245 pKa = 6.93HH246 pKa = 5.96SVEE249 pKa = 4.0DD250 pKa = 3.55RR251 pKa = 11.84FFQEE255 pKa = 3.47WVEE258 pKa = 3.99THH260 pKa = 7.34DD261 pKa = 4.07IDD263 pKa = 6.38LEE265 pKa = 4.29VSDD268 pKa = 5.63CNLDD272 pKa = 3.29MSVFNDD278 pKa = 3.23WSKK281 pKa = 10.37GTDD284 pKa = 3.19TRR286 pKa = 11.84LVPQMCVGGLSHH298 pKa = 7.2RR299 pKa = 11.84VPTQRR304 pKa = 11.84EE305 pKa = 3.62ALLAIKK311 pKa = 9.37KK312 pKa = 10.3RR313 pKa = 11.84NMNVPEE319 pKa = 4.35LQSQFDD325 pKa = 3.99HH326 pKa = 6.91EE327 pKa = 5.03DD328 pKa = 3.34VLNRR332 pKa = 11.84CVNRR336 pKa = 11.84FLTHH340 pKa = 6.03VVDD343 pKa = 3.8KK344 pKa = 10.85TRR346 pKa = 11.84LGKK349 pKa = 10.54LMPISGEE356 pKa = 3.65EE357 pKa = 4.14VYY359 pKa = 10.74FFNQYY364 pKa = 10.71LEE366 pKa = 4.39NKK368 pKa = 9.44NPPLSEE374 pKa = 4.1YY375 pKa = 10.32KK376 pKa = 10.97GPVPLLALDD385 pKa = 4.8KK386 pKa = 10.0YY387 pKa = 9.42MHH389 pKa = 6.63MIKK392 pKa = 7.75TTLKK396 pKa = 10.23PVEE399 pKa = 4.94DD400 pKa = 4.74DD401 pKa = 3.36SLHH404 pKa = 6.45IEE406 pKa = 4.21RR407 pKa = 11.84PVPATITYY415 pKa = 8.29HH416 pKa = 5.88KK417 pKa = 9.69KK418 pKa = 9.51GVVMMTSPYY427 pKa = 9.61FLCAMVRR434 pKa = 11.84LLYY437 pKa = 10.17VLKK440 pKa = 10.61SKK442 pKa = 10.77VFIPTGKK449 pKa = 9.24YY450 pKa = 9.14HH451 pKa = 7.24QIFQMDD457 pKa = 4.13PVRR460 pKa = 11.84LKK462 pKa = 10.4NSKK465 pKa = 9.6FFKK468 pKa = 10.62EE469 pKa = 3.42IDD471 pKa = 3.37FSKK474 pKa = 10.51FDD476 pKa = 3.37KK477 pKa = 11.04SQGRR481 pKa = 11.84LHH483 pKa = 7.08HH484 pKa = 7.0DD485 pKa = 3.21VQLRR489 pKa = 11.84LFLHH493 pKa = 6.66LGIPQHH499 pKa = 6.96FVDD502 pKa = 3.62TWFNAHH508 pKa = 5.96EE509 pKa = 4.3RR510 pKa = 11.84SHH512 pKa = 7.94IRR514 pKa = 11.84DD515 pKa = 3.64RR516 pKa = 11.84DD517 pKa = 3.73CGIGFSVDD525 pKa = 3.25YY526 pKa = 10.46QRR528 pKa = 11.84RR529 pKa = 11.84TGDD532 pKa = 2.74ACTYY536 pKa = 10.46LGNTLVTLSVLSYY549 pKa = 11.1VYY551 pKa = 10.62DD552 pKa = 3.92LSKK555 pKa = 10.95PDD557 pKa = 3.62ILFVAASGDD566 pKa = 3.57DD567 pKa = 3.75SLIGSLNPLPRR578 pKa = 11.84DD579 pKa = 3.69KK580 pKa = 11.33EE581 pKa = 4.21DD582 pKa = 3.51LCVSLFNLEE591 pKa = 3.88TKK593 pKa = 10.47FPHH596 pKa = 5.74NQPFLCSKK604 pKa = 9.95FLLVVEE610 pKa = 5.04CDD612 pKa = 3.71DD613 pKa = 3.73GTEE616 pKa = 3.98EE617 pKa = 4.32VLAVPNPLKK626 pKa = 10.32ILQKK630 pKa = 9.95MGPKK634 pKa = 9.47NLQVTVLDD642 pKa = 4.97DD643 pKa = 4.31YY644 pKa = 11.44YY645 pKa = 11.6QSLCDD650 pKa = 3.68ILWVFEE656 pKa = 4.54DD657 pKa = 3.8ADD659 pKa = 3.3ICRR662 pKa = 11.84RR663 pKa = 11.84VAQLAEE669 pKa = 3.72YY670 pKa = 10.1RR671 pKa = 11.84QFKK674 pKa = 9.91GKK676 pKa = 9.92RR677 pKa = 11.84SCVFLEE683 pKa = 4.11SALLSLPSLVANRR696 pKa = 11.84LKK698 pKa = 10.85FIRR701 pKa = 11.84RR702 pKa = 11.84TINLEE707 pKa = 3.6GSKK710 pKa = 10.85ACIRR714 pKa = 11.84NDD716 pKa = 3.11AYY718 pKa = 11.75SNLIACNVSRR728 pKa = 11.84AKK730 pKa = 10.32CGRR733 pKa = 11.84DD734 pKa = 3.08AAGSDD739 pKa = 3.75SVNSARR745 pKa = 11.84EE746 pKa = 4.1SVPTRR751 pKa = 11.84TPKK754 pKa = 10.18EE755 pKa = 3.72WGGSRR760 pKa = 11.84KK761 pKa = 9.68RR762 pKa = 11.84NPGCAVRR769 pKa = 11.84RR770 pKa = 11.84EE771 pKa = 4.29GVRR774 pKa = 11.84EE775 pKa = 3.78GDD777 pKa = 2.8RR778 pKa = 11.84SEE780 pKa = 4.46FNGTGSHH787 pKa = 5.17RR788 pKa = 11.84TDD790 pKa = 3.39YY791 pKa = 11.04GRR793 pKa = 11.84KK794 pKa = 8.39KK795 pKa = 9.99PSRR798 pKa = 11.84KK799 pKa = 10.01GGFKK803 pKa = 10.02VHH805 pKa = 6.7
MM1 pKa = 7.42DD2 pKa = 4.85QIRR5 pKa = 11.84INLIDD10 pKa = 3.85SFLSSRR16 pKa = 11.84LCFNATFGVYY26 pKa = 9.97HH27 pKa = 7.73DD28 pKa = 6.31DD29 pKa = 3.44YY30 pKa = 10.91MEE32 pKa = 4.05WVKK35 pKa = 10.95LFLFKK40 pKa = 10.7HH41 pKa = 5.64VIEE44 pKa = 4.61HH45 pKa = 5.19TAKK48 pKa = 10.22FDD50 pKa = 3.39FATIHH55 pKa = 5.5MTLEE59 pKa = 4.03MVLCRR64 pKa = 11.84LVPGLGDD71 pKa = 3.31ALYY74 pKa = 9.94PEE76 pKa = 5.17EE77 pKa = 4.19EE78 pKa = 4.37NEE80 pKa = 4.03VDD82 pKa = 3.52GLSSRR87 pKa = 11.84EE88 pKa = 3.48IDD90 pKa = 3.94PFYY93 pKa = 11.13LPYY96 pKa = 10.75DD97 pKa = 4.42DD98 pKa = 6.49LDD100 pKa = 3.52VDD102 pKa = 3.87YY103 pKa = 9.26TALCRR108 pKa = 11.84EE109 pKa = 4.42RR110 pKa = 11.84DD111 pKa = 3.71DD112 pKa = 5.27SIPTIPPEE120 pKa = 4.47DD121 pKa = 4.05EE122 pKa = 4.25LSVIVKK128 pKa = 10.05NMTYY132 pKa = 9.91IPEE135 pKa = 4.38GTSWGDD141 pKa = 3.23VSDD144 pKa = 5.25SSFQEE149 pKa = 4.01HH150 pKa = 6.79LEE152 pKa = 4.34EE153 pKa = 4.35IQPVPLGSDD162 pKa = 3.25VGHH165 pKa = 7.18TIPNEE170 pKa = 4.31HH171 pKa = 7.05IPFDD175 pKa = 5.07DD176 pKa = 5.19DD177 pKa = 3.8GQLVDD182 pKa = 4.54VQWSDD187 pKa = 4.77AIPSLATDD195 pKa = 3.85VTCDD199 pKa = 3.04ADD201 pKa = 3.62YY202 pKa = 10.99RR203 pKa = 11.84EE204 pKa = 4.92CGFVSYY210 pKa = 11.05SSGSQHH216 pKa = 6.44SSRR219 pKa = 11.84WKK221 pKa = 9.83PKK223 pKa = 9.11VSQVKK228 pKa = 9.2PDD230 pKa = 3.64PSIIQSAIDD239 pKa = 3.46EE240 pKa = 5.0LFPHH244 pKa = 6.27HH245 pKa = 6.93HH246 pKa = 5.96SVEE249 pKa = 4.0DD250 pKa = 3.55RR251 pKa = 11.84FFQEE255 pKa = 3.47WVEE258 pKa = 3.99THH260 pKa = 7.34DD261 pKa = 4.07IDD263 pKa = 6.38LEE265 pKa = 4.29VSDD268 pKa = 5.63CNLDD272 pKa = 3.29MSVFNDD278 pKa = 3.23WSKK281 pKa = 10.37GTDD284 pKa = 3.19TRR286 pKa = 11.84LVPQMCVGGLSHH298 pKa = 7.2RR299 pKa = 11.84VPTQRR304 pKa = 11.84EE305 pKa = 3.62ALLAIKK311 pKa = 9.37KK312 pKa = 10.3RR313 pKa = 11.84NMNVPEE319 pKa = 4.35LQSQFDD325 pKa = 3.99HH326 pKa = 6.91EE327 pKa = 5.03DD328 pKa = 3.34VLNRR332 pKa = 11.84CVNRR336 pKa = 11.84FLTHH340 pKa = 6.03VVDD343 pKa = 3.8KK344 pKa = 10.85TRR346 pKa = 11.84LGKK349 pKa = 10.54LMPISGEE356 pKa = 3.65EE357 pKa = 4.14VYY359 pKa = 10.74FFNQYY364 pKa = 10.71LEE366 pKa = 4.39NKK368 pKa = 9.44NPPLSEE374 pKa = 4.1YY375 pKa = 10.32KK376 pKa = 10.97GPVPLLALDD385 pKa = 4.8KK386 pKa = 10.0YY387 pKa = 9.42MHH389 pKa = 6.63MIKK392 pKa = 7.75TTLKK396 pKa = 10.23PVEE399 pKa = 4.94DD400 pKa = 4.74DD401 pKa = 3.36SLHH404 pKa = 6.45IEE406 pKa = 4.21RR407 pKa = 11.84PVPATITYY415 pKa = 8.29HH416 pKa = 5.88KK417 pKa = 9.69KK418 pKa = 9.51GVVMMTSPYY427 pKa = 9.61FLCAMVRR434 pKa = 11.84LLYY437 pKa = 10.17VLKK440 pKa = 10.61SKK442 pKa = 10.77VFIPTGKK449 pKa = 9.24YY450 pKa = 9.14HH451 pKa = 7.24QIFQMDD457 pKa = 4.13PVRR460 pKa = 11.84LKK462 pKa = 10.4NSKK465 pKa = 9.6FFKK468 pKa = 10.62EE469 pKa = 3.42IDD471 pKa = 3.37FSKK474 pKa = 10.51FDD476 pKa = 3.37KK477 pKa = 11.04SQGRR481 pKa = 11.84LHH483 pKa = 7.08HH484 pKa = 7.0DD485 pKa = 3.21VQLRR489 pKa = 11.84LFLHH493 pKa = 6.66LGIPQHH499 pKa = 6.96FVDD502 pKa = 3.62TWFNAHH508 pKa = 5.96EE509 pKa = 4.3RR510 pKa = 11.84SHH512 pKa = 7.94IRR514 pKa = 11.84DD515 pKa = 3.64RR516 pKa = 11.84DD517 pKa = 3.73CGIGFSVDD525 pKa = 3.25YY526 pKa = 10.46QRR528 pKa = 11.84RR529 pKa = 11.84TGDD532 pKa = 2.74ACTYY536 pKa = 10.46LGNTLVTLSVLSYY549 pKa = 11.1VYY551 pKa = 10.62DD552 pKa = 3.92LSKK555 pKa = 10.95PDD557 pKa = 3.62ILFVAASGDD566 pKa = 3.57DD567 pKa = 3.75SLIGSLNPLPRR578 pKa = 11.84DD579 pKa = 3.69KK580 pKa = 11.33EE581 pKa = 4.21DD582 pKa = 3.51LCVSLFNLEE591 pKa = 3.88TKK593 pKa = 10.47FPHH596 pKa = 5.74NQPFLCSKK604 pKa = 9.95FLLVVEE610 pKa = 5.04CDD612 pKa = 3.71DD613 pKa = 3.73GTEE616 pKa = 3.98EE617 pKa = 4.32VLAVPNPLKK626 pKa = 10.32ILQKK630 pKa = 9.95MGPKK634 pKa = 9.47NLQVTVLDD642 pKa = 4.97DD643 pKa = 4.31YY644 pKa = 11.44YY645 pKa = 11.6QSLCDD650 pKa = 3.68ILWVFEE656 pKa = 4.54DD657 pKa = 3.8ADD659 pKa = 3.3ICRR662 pKa = 11.84RR663 pKa = 11.84VAQLAEE669 pKa = 3.72YY670 pKa = 10.1RR671 pKa = 11.84QFKK674 pKa = 9.91GKK676 pKa = 9.92RR677 pKa = 11.84SCVFLEE683 pKa = 4.11SALLSLPSLVANRR696 pKa = 11.84LKK698 pKa = 10.85FIRR701 pKa = 11.84RR702 pKa = 11.84TINLEE707 pKa = 3.6GSKK710 pKa = 10.85ACIRR714 pKa = 11.84NDD716 pKa = 3.11AYY718 pKa = 11.75SNLIACNVSRR728 pKa = 11.84AKK730 pKa = 10.32CGRR733 pKa = 11.84DD734 pKa = 3.08AAGSDD739 pKa = 3.75SVNSARR745 pKa = 11.84EE746 pKa = 4.1SVPTRR751 pKa = 11.84TPKK754 pKa = 10.18EE755 pKa = 3.72WGGSRR760 pKa = 11.84KK761 pKa = 9.68RR762 pKa = 11.84NPGCAVRR769 pKa = 11.84RR770 pKa = 11.84EE771 pKa = 4.29GVRR774 pKa = 11.84EE775 pKa = 3.78GDD777 pKa = 2.8RR778 pKa = 11.84SEE780 pKa = 4.46FNGTGSHH787 pKa = 5.17RR788 pKa = 11.84TDD790 pKa = 3.39YY791 pKa = 11.04GRR793 pKa = 11.84KK794 pKa = 8.39KK795 pKa = 9.99PSRR798 pKa = 11.84KK799 pKa = 10.01GGFKK803 pKa = 10.02VHH805 pKa = 6.7
Molecular weight: 91.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q80852|Q80852_9BROM Coat protein OS=Parietaria mottle virus OX=64958 PE=2 SV=1
MM1 pKa = 7.45YY2 pKa = 10.39EE3 pKa = 4.52LFNHH7 pKa = 6.35SLQPNQLTPCLRR19 pKa = 11.84TRR21 pKa = 11.84ANGGQVSNRR30 pKa = 11.84QSRR33 pKa = 11.84NVRR36 pKa = 11.84RR37 pKa = 11.84AAAFRR42 pKa = 11.84NSQQTSARR50 pKa = 11.84IPVPVPVIPVSRR62 pKa = 11.84PNGPKK67 pKa = 10.33ASLKK71 pKa = 10.67LPNNQVWVYY80 pKa = 10.62KK81 pKa = 10.37VASEE85 pKa = 4.44LAAKK89 pKa = 9.78TSDD92 pKa = 3.49ANDD95 pKa = 4.76AISLTTMLSGISDD108 pKa = 3.72VKK110 pKa = 11.04PEE112 pKa = 3.98TKK114 pKa = 10.33LYY116 pKa = 10.04RR117 pKa = 11.84LVFGFVAEE125 pKa = 4.28SDD127 pKa = 3.67GSFGVVEE134 pKa = 4.69DD135 pKa = 4.63EE136 pKa = 4.18NVSGNVVPDD145 pKa = 3.81PPVVGRR151 pKa = 11.84AGFKK155 pKa = 10.29KK156 pKa = 8.89HH157 pKa = 5.32TNKK160 pKa = 9.86CRR162 pKa = 11.84DD163 pKa = 3.22VNLEE167 pKa = 3.75GKK169 pKa = 8.16TPDD172 pKa = 3.48EE173 pKa = 4.77LKK175 pKa = 11.15NMAVVWCLDD184 pKa = 3.57EE185 pKa = 4.02NRR187 pKa = 11.84KK188 pKa = 7.52VAKK191 pKa = 10.2RR192 pKa = 11.84IAFTHH197 pKa = 6.0FWFAISRR204 pKa = 11.84PSPLMPPEE212 pKa = 4.78NILVDD217 pKa = 4.26GNQQ220 pKa = 2.93
MM1 pKa = 7.45YY2 pKa = 10.39EE3 pKa = 4.52LFNHH7 pKa = 6.35SLQPNQLTPCLRR19 pKa = 11.84TRR21 pKa = 11.84ANGGQVSNRR30 pKa = 11.84QSRR33 pKa = 11.84NVRR36 pKa = 11.84RR37 pKa = 11.84AAAFRR42 pKa = 11.84NSQQTSARR50 pKa = 11.84IPVPVPVIPVSRR62 pKa = 11.84PNGPKK67 pKa = 10.33ASLKK71 pKa = 10.67LPNNQVWVYY80 pKa = 10.62KK81 pKa = 10.37VASEE85 pKa = 4.44LAAKK89 pKa = 9.78TSDD92 pKa = 3.49ANDD95 pKa = 4.76AISLTTMLSGISDD108 pKa = 3.72VKK110 pKa = 11.04PEE112 pKa = 3.98TKK114 pKa = 10.33LYY116 pKa = 10.04RR117 pKa = 11.84LVFGFVAEE125 pKa = 4.28SDD127 pKa = 3.67GSFGVVEE134 pKa = 4.69DD135 pKa = 4.63EE136 pKa = 4.18NVSGNVVPDD145 pKa = 3.81PPVVGRR151 pKa = 11.84AGFKK155 pKa = 10.29KK156 pKa = 8.89HH157 pKa = 5.32TNKK160 pKa = 9.86CRR162 pKa = 11.84DD163 pKa = 3.22VNLEE167 pKa = 3.75GKK169 pKa = 8.16TPDD172 pKa = 3.48EE173 pKa = 4.77LKK175 pKa = 11.15NMAVVWCLDD184 pKa = 3.57EE185 pKa = 4.02NRR187 pKa = 11.84KK188 pKa = 7.52VAKK191 pKa = 10.2RR192 pKa = 11.84IAFTHH197 pKa = 6.0FWFAISRR204 pKa = 11.84PSPLMPPEE212 pKa = 4.78NILVDD217 pKa = 4.26GNQQ220 pKa = 2.93
Molecular weight: 24.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2621 |
205 |
1098 |
524.2 |
58.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.066 ± 0.855 | 2.251 ± 0.215 |
7.02 ± 0.682 | 5.799 ± 0.185 |
4.273 ± 0.335 | 4.922 ± 0.424 |
2.633 ± 0.428 | 5.151 ± 0.439 |
6.219 ± 0.337 | 9.271 ± 0.366 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.167 | 4.388 ± 0.568 |
5.151 ± 0.918 | 2.671 ± 0.338 |
5.799 ± 0.246 | 8.394 ± 0.294 |
5.799 ± 0.766 | 8.127 ± 0.445 |
1.03 ± 0.078 | 2.709 ± 0.539 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
