
Grosmannia clavigera (strain kw1407 / UAMH 11150) (Blue stain fungus) (Graphiocladiella clavigera)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Ophiostomatales; Ophiostomataceae; Grosmannia; Grosmannia clavigera
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
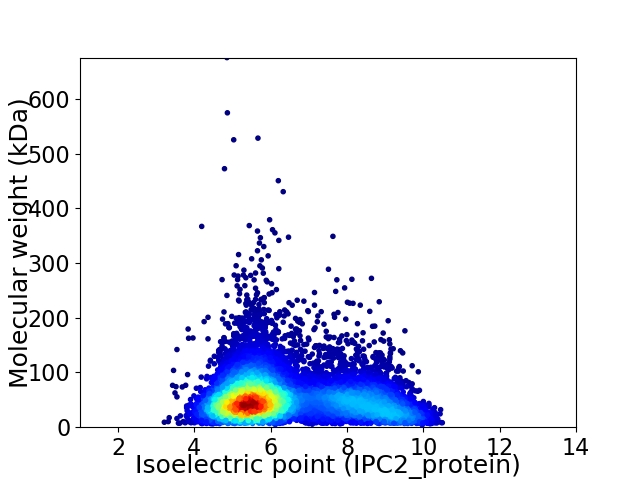
Virtual 2D-PAGE plot for 8311 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0XL67|F0XL67_GROCL Amino acid permease OS=Grosmannia clavigera (strain kw1407 / UAMH 11150) OX=655863 GN=CMQ_8220 PE=4 SV=1
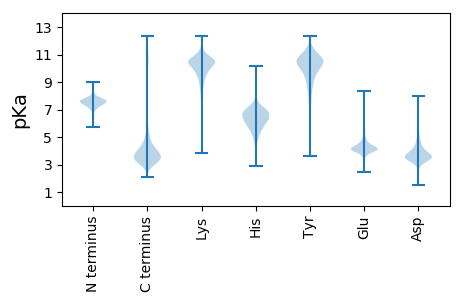
MM1 pKa = 7.16RR2 pKa = 11.84QEE4 pKa = 3.85IGEE7 pKa = 4.38GLVDD11 pKa = 3.79TPLVEE16 pKa = 4.24TASQLVQGGGVAGDD30 pKa = 3.44VFVRR34 pKa = 11.84SLLALWRR41 pKa = 11.84IAVVADD47 pKa = 4.8FSSVAITAPVSDD59 pKa = 4.79DD60 pKa = 3.51VASAEE65 pKa = 3.68IWCFVFCGLRR75 pKa = 11.84LIYY78 pKa = 10.28YY79 pKa = 8.94GVVGLINDD87 pKa = 4.76GYY89 pKa = 10.47DD90 pKa = 3.1KK91 pKa = 10.97KK92 pKa = 11.05FKK94 pKa = 10.85VYY96 pKa = 10.64AKK98 pKa = 10.03VVAGGADD105 pKa = 3.73LQVHH109 pKa = 7.17DD110 pKa = 5.4FMQQIGDD117 pKa = 4.03KK118 pKa = 10.89DD119 pKa = 3.97SLDD122 pKa = 3.61KK123 pKa = 11.36SDD125 pKa = 5.32GYY127 pKa = 10.23NTMANLALNMSFASEE142 pKa = 4.48SEE144 pKa = 4.07PCPGDD149 pKa = 3.38FSEE152 pKa = 5.33RR153 pKa = 11.84GYY155 pKa = 11.61GPDD158 pKa = 3.43CLYY161 pKa = 10.84EE162 pKa = 3.88QTVYY166 pKa = 7.72WTLRR170 pKa = 11.84DD171 pKa = 3.68DD172 pKa = 3.9QADD175 pKa = 3.41VFYY178 pKa = 11.29ANLEE182 pKa = 4.27SATGIPKK189 pKa = 10.43DD190 pKa = 3.77KK191 pKa = 10.74IGFGYY196 pKa = 8.57YY197 pKa = 9.73TDD199 pKa = 4.31IDD201 pKa = 3.84NCSGSGSKK209 pKa = 10.61VGDD212 pKa = 4.53GDD214 pKa = 3.86NCWNIDD220 pKa = 3.78YY221 pKa = 10.78EE222 pKa = 4.6LGAPFPNGYY231 pKa = 8.28GTDD234 pKa = 3.72DD235 pKa = 3.46VANPKK240 pKa = 10.11DD241 pKa = 3.55VAKK244 pKa = 10.57II245 pKa = 3.63
MM1 pKa = 7.16RR2 pKa = 11.84QEE4 pKa = 3.85IGEE7 pKa = 4.38GLVDD11 pKa = 3.79TPLVEE16 pKa = 4.24TASQLVQGGGVAGDD30 pKa = 3.44VFVRR34 pKa = 11.84SLLALWRR41 pKa = 11.84IAVVADD47 pKa = 4.8FSSVAITAPVSDD59 pKa = 4.79DD60 pKa = 3.51VASAEE65 pKa = 3.68IWCFVFCGLRR75 pKa = 11.84LIYY78 pKa = 10.28YY79 pKa = 8.94GVVGLINDD87 pKa = 4.76GYY89 pKa = 10.47DD90 pKa = 3.1KK91 pKa = 10.97KK92 pKa = 11.05FKK94 pKa = 10.85VYY96 pKa = 10.64AKK98 pKa = 10.03VVAGGADD105 pKa = 3.73LQVHH109 pKa = 7.17DD110 pKa = 5.4FMQQIGDD117 pKa = 4.03KK118 pKa = 10.89DD119 pKa = 3.97SLDD122 pKa = 3.61KK123 pKa = 11.36SDD125 pKa = 5.32GYY127 pKa = 10.23NTMANLALNMSFASEE142 pKa = 4.48SEE144 pKa = 4.07PCPGDD149 pKa = 3.38FSEE152 pKa = 5.33RR153 pKa = 11.84GYY155 pKa = 11.61GPDD158 pKa = 3.43CLYY161 pKa = 10.84EE162 pKa = 3.88QTVYY166 pKa = 7.72WTLRR170 pKa = 11.84DD171 pKa = 3.68DD172 pKa = 3.9QADD175 pKa = 3.41VFYY178 pKa = 11.29ANLEE182 pKa = 4.27SATGIPKK189 pKa = 10.43DD190 pKa = 3.77KK191 pKa = 10.74IGFGYY196 pKa = 8.57YY197 pKa = 9.73TDD199 pKa = 4.31IDD201 pKa = 3.84NCSGSGSKK209 pKa = 10.61VGDD212 pKa = 4.53GDD214 pKa = 3.86NCWNIDD220 pKa = 3.78YY221 pKa = 10.78EE222 pKa = 4.6LGAPFPNGYY231 pKa = 8.28GTDD234 pKa = 3.72DD235 pKa = 3.46VANPKK240 pKa = 10.11DD241 pKa = 3.55VAKK244 pKa = 10.57II245 pKa = 3.63
Molecular weight: 26.47 kDa
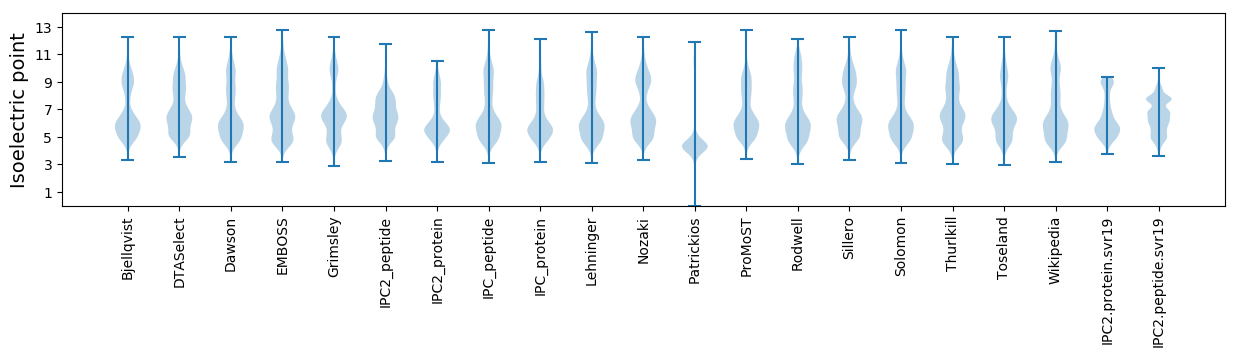
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0X9S3|F0X9S3_GROCL Benzoate 4-monooxygenase cytochrome p450 OS=Grosmannia clavigera (strain kw1407 / UAMH 11150) OX=655863 GN=CMQ_3395 PE=3 SV=1
MM1 pKa = 7.02QSAYY5 pKa = 10.72SAFSLAVPFPCGQAATHH22 pKa = 6.06AVVAFTCAALGSGACDD38 pKa = 3.59GQSMLLALGWKK49 pKa = 9.25KK50 pKa = 9.12RR51 pKa = 11.84TYY53 pKa = 11.32GNTLGPARR61 pKa = 11.84DD62 pKa = 3.61VARR65 pKa = 11.84IAGSEE70 pKa = 3.76ARR72 pKa = 11.84GDD74 pKa = 3.52GRR76 pKa = 11.84LVRR79 pKa = 11.84GLGRR83 pKa = 11.84AEE85 pKa = 3.57AVGRR89 pKa = 11.84RR90 pKa = 11.84GHH92 pKa = 6.38KK93 pKa = 10.21GSGSRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84GGLGRR106 pKa = 11.84GKK108 pKa = 10.46GSKK111 pKa = 9.8EE112 pKa = 3.39ADD114 pKa = 2.93EE115 pKa = 4.98SGRR118 pKa = 11.84EE119 pKa = 3.86THH121 pKa = 6.82GDD123 pKa = 3.65GMVLWTRR130 pKa = 11.84GVYY133 pKa = 10.41CILLEE138 pKa = 4.16RR139 pKa = 4.92
MM1 pKa = 7.02QSAYY5 pKa = 10.72SAFSLAVPFPCGQAATHH22 pKa = 6.06AVVAFTCAALGSGACDD38 pKa = 3.59GQSMLLALGWKK49 pKa = 9.25KK50 pKa = 9.12RR51 pKa = 11.84TYY53 pKa = 11.32GNTLGPARR61 pKa = 11.84DD62 pKa = 3.61VARR65 pKa = 11.84IAGSEE70 pKa = 3.76ARR72 pKa = 11.84GDD74 pKa = 3.52GRR76 pKa = 11.84LVRR79 pKa = 11.84GLGRR83 pKa = 11.84AEE85 pKa = 3.57AVGRR89 pKa = 11.84RR90 pKa = 11.84GHH92 pKa = 6.38KK93 pKa = 10.21GSGSRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84GGLGRR106 pKa = 11.84GKK108 pKa = 10.46GSKK111 pKa = 9.8EE112 pKa = 3.39ADD114 pKa = 2.93EE115 pKa = 4.98SGRR118 pKa = 11.84EE119 pKa = 3.86THH121 pKa = 6.82GDD123 pKa = 3.65GMVLWTRR130 pKa = 11.84GVYY133 pKa = 10.41CILLEE138 pKa = 4.16RR139 pKa = 4.92
Molecular weight: 14.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4531634 |
35 |
6207 |
545.3 |
59.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.425 ± 0.03 | 1.156 ± 0.009 |
5.921 ± 0.017 | 5.716 ± 0.026 |
3.408 ± 0.016 | 7.414 ± 0.021 |
2.367 ± 0.011 | 4.08 ± 0.016 |
3.918 ± 0.023 | 8.906 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.148 ± 0.009 | 3.13 ± 0.014 |
5.904 ± 0.025 | 4.023 ± 0.02 |
6.669 ± 0.025 | 8.318 ± 0.033 |
6.053 ± 0.018 | 6.577 ± 0.021 |
1.352 ± 0.009 | 2.516 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
