
Echinicola vietnamensis (strain DSM 17526 / LMG 23754 / KMM 6221)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Echinicola; Echinicola vietnamensis
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
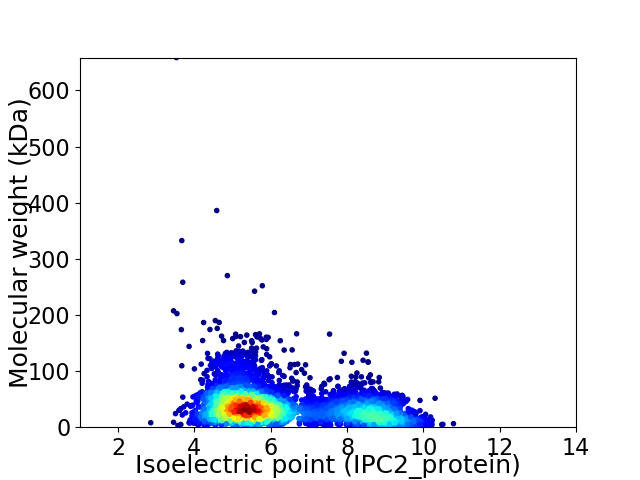
Virtual 2D-PAGE plot for 4509 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
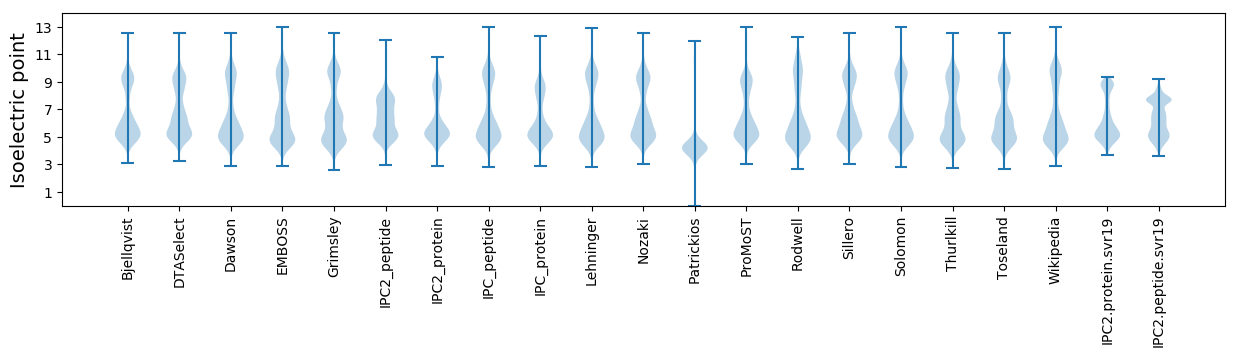
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0FUQ2|L0FUQ2_ECHVK Uncharacterized protein OS=Echinicola vietnamensis (strain DSM 17526 / LMG 23754 / KMM 6221) OX=926556 GN=Echvi_1350 PE=4 SV=1
MM1 pKa = 7.48HH2 pKa = 7.69KK3 pKa = 10.05ISYY6 pKa = 10.1ISILVVCAILVGYY19 pKa = 8.41VPKK22 pKa = 10.79DD23 pKa = 3.47VVSNSGVKK31 pKa = 10.1VEE33 pKa = 4.52SVRR36 pKa = 11.84KK37 pKa = 9.85NINSDD42 pKa = 2.66TDD44 pKa = 3.62YY45 pKa = 11.86DD46 pKa = 4.05EE47 pKa = 6.96DD48 pKa = 5.34GVLNDD53 pKa = 5.25NDD55 pKa = 5.49LDD57 pKa = 4.43DD58 pKa = 6.26DD59 pKa = 4.41NDD61 pKa = 4.89GILDD65 pKa = 3.44IDD67 pKa = 3.85EE68 pKa = 5.61GFVCTTNSQKK78 pKa = 9.51TVLKK82 pKa = 10.39AISNNIPAEE91 pKa = 4.18GTSYY95 pKa = 11.14CGGLKK100 pKa = 10.28NSYY103 pKa = 7.4THH105 pKa = 7.2FSDD108 pKa = 6.02GYY110 pKa = 11.1LDD112 pKa = 3.73TQNDD116 pKa = 3.07ITGTYY121 pKa = 7.87TLNYY125 pKa = 9.1SYY127 pKa = 11.37SGLDD131 pKa = 3.29VSKK134 pKa = 10.75EE135 pKa = 3.87AIFQHH140 pKa = 6.17RR141 pKa = 11.84LVYY144 pKa = 10.83SNVNGSHH151 pKa = 6.5GVDD154 pKa = 2.97QNTSNVRR161 pKa = 11.84ITSKK165 pKa = 10.35IYY167 pKa = 10.73VNNTLVRR174 pKa = 11.84TASVTGWNYY183 pKa = 10.09HH184 pKa = 4.5SQPRR188 pKa = 11.84EE189 pKa = 3.67KK190 pKa = 10.7DD191 pKa = 2.89NKK193 pKa = 10.02YY194 pKa = 11.09YY195 pKa = 11.05YY196 pKa = 7.28FTPSTASGTVRR207 pKa = 11.84IDD209 pKa = 3.06ITFEE213 pKa = 3.96RR214 pKa = 11.84LNGYY218 pKa = 9.89CDD220 pKa = 3.98VIPEE224 pKa = 4.18VFFGGNLSQTSDD236 pKa = 4.79PICTSIDD243 pKa = 3.37TDD245 pKa = 3.69GDD247 pKa = 4.4GIPDD251 pKa = 3.97HH252 pKa = 7.11QDD254 pKa = 2.93LDD256 pKa = 4.2SDD258 pKa = 4.88GDD260 pKa = 3.95GCPDD264 pKa = 3.36ALEE267 pKa = 4.51GQGGFTAADD276 pKa = 3.81LVSSALAGGNSGANYY291 pKa = 8.97TGVAGSVVSNLGATVGSNGVPTKK314 pKa = 10.41AASGQGRR321 pKa = 11.84GTSRR325 pKa = 11.84EE326 pKa = 3.96IQQVASACIGCSTLAGDD343 pKa = 3.97VTDD346 pKa = 3.92TGGDD350 pKa = 3.53PVGNAMIRR358 pKa = 11.84VYY360 pKa = 11.41ADD362 pKa = 3.61FDD364 pKa = 4.14QNGNISSGDD373 pKa = 3.52EE374 pKa = 3.75LLKK377 pKa = 10.93VVNADD382 pKa = 3.09AQGAYY387 pKa = 10.14SINKK391 pKa = 9.24GYY393 pKa = 10.69YY394 pKa = 8.11ITVKK398 pKa = 10.67DD399 pKa = 4.45DD400 pKa = 3.65FSGDD404 pKa = 3.47VFSGNNNSSGDD415 pKa = 3.59YY416 pKa = 10.27NAWLDD421 pKa = 4.39ASWTQSNSANSGISSNTNGDD441 pKa = 3.73DD442 pKa = 3.73NLEE445 pKa = 3.98SRR447 pKa = 11.84HH448 pKa = 6.6IYY450 pKa = 8.77IHH452 pKa = 6.74NNTQVYY458 pKa = 9.58RR459 pKa = 11.84AVNLSKK465 pKa = 10.91FKK467 pKa = 9.86TANLRR472 pKa = 11.84FDD474 pKa = 4.25YY475 pKa = 9.39DD476 pKa = 3.77TEE478 pKa = 4.59NDD480 pKa = 3.58LDD482 pKa = 3.93ATDD485 pKa = 4.0KK486 pKa = 11.19FHH488 pKa = 8.01VEE490 pKa = 3.84LSKK493 pKa = 11.44DD494 pKa = 3.12GGATYY499 pKa = 7.68TTVYY503 pKa = 9.93TYY505 pKa = 11.24TDD507 pKa = 3.81GNVDD511 pKa = 3.54NQPARR516 pKa = 11.84SQNINLNGYY525 pKa = 9.3AGEE528 pKa = 4.29SNVIIRR534 pKa = 11.84FRR536 pKa = 11.84AEE538 pKa = 3.96SSSGEE543 pKa = 4.37YY544 pKa = 9.61IWLDD548 pKa = 3.01NVTIYY553 pKa = 11.15NAFPTVLLKK562 pKa = 10.24PVYY565 pKa = 9.43NEE567 pKa = 4.58PYY569 pKa = 9.83RR570 pKa = 11.84SSNPATIAVNPSTTTCSGLDD590 pKa = 3.52FTLLDD595 pKa = 4.26ACDD598 pKa = 3.99VDD600 pKa = 4.16PTDD603 pKa = 3.32TDD605 pKa = 3.72GDD607 pKa = 4.72GVCDD611 pKa = 6.08AIDD614 pKa = 4.72IDD616 pKa = 5.27DD617 pKa = 5.49DD618 pKa = 4.87DD619 pKa = 7.17DD620 pKa = 4.5GVLDD624 pKa = 3.58EE625 pKa = 5.13TEE627 pKa = 4.05GHH629 pKa = 6.4FCGEE633 pKa = 4.27LDD635 pKa = 3.51RR636 pKa = 11.84NIIIGYY642 pKa = 9.19LDD644 pKa = 3.55SGVGNDD650 pKa = 4.51GLATDD655 pKa = 4.21MLLNLKK661 pKa = 10.4NYY663 pKa = 10.18GEE665 pKa = 4.45YY666 pKa = 9.54GTYY669 pKa = 10.54DD670 pKa = 3.13KK671 pKa = 11.44VRR673 pKa = 11.84GVVLVPFSSSAEE685 pKa = 3.67ITEE688 pKa = 4.71ANLEE692 pKa = 4.17ANNIDD697 pKa = 3.65VFFAGSSTANSEE709 pKa = 3.98GSSDD713 pKa = 3.79KK714 pKa = 11.12LSSSTNAVLMSWAEE728 pKa = 4.04NNDD731 pKa = 2.96KK732 pKa = 11.27GLFVLQNNAVDD743 pKa = 3.55YY744 pKa = 10.85GYY746 pKa = 8.48TLTNNDD752 pKa = 4.8LPPDD756 pKa = 3.83VPNGEE761 pKa = 4.71LGYY764 pKa = 10.22QVYY767 pKa = 9.63TNGYY771 pKa = 8.24WPVSSITQTGTVKK784 pKa = 8.23MTISSSTRR792 pKa = 11.84SFEE795 pKa = 4.27ALMVDD800 pKa = 3.84SKK802 pKa = 11.61SRR804 pKa = 11.84PTLVKK809 pKa = 10.73DD810 pKa = 3.64NMMKK814 pKa = 10.64LVIFPDD820 pKa = 3.13ATIYY824 pKa = 10.87NDD826 pKa = 3.38EE827 pKa = 4.89ANTVDD832 pKa = 4.21PNKK835 pKa = 10.66NDD837 pKa = 3.37ATKK840 pKa = 10.84VIADD844 pKa = 3.79TWAFVMDD851 pKa = 3.88TFIGDD856 pKa = 3.72GCSEE860 pKa = 5.02LDD862 pKa = 3.67TNGDD866 pKa = 3.54GTPNHH871 pKa = 7.18LDD873 pKa = 3.69LDD875 pKa = 4.25SDD877 pKa = 4.55GDD879 pKa = 3.99GCSDD883 pKa = 3.48AFEE886 pKa = 5.01ANATEE891 pKa = 5.7DD892 pKa = 3.3ITPNFQFDD900 pKa = 3.97VQKK903 pKa = 10.22GTSSDD908 pKa = 3.62TNGDD912 pKa = 3.62GLADD916 pKa = 3.93AVDD919 pKa = 3.97EE920 pKa = 4.43NLNGIPDD927 pKa = 4.14YY928 pKa = 11.42VSAYY932 pKa = 10.65YY933 pKa = 9.89PDD935 pKa = 5.11ALDD938 pKa = 3.75NTTCTDD944 pKa = 3.12SDD946 pKa = 3.74GDD948 pKa = 4.34TIVDD952 pKa = 4.73AVDD955 pKa = 5.13LDD957 pKa = 4.27DD958 pKa = 6.61DD959 pKa = 4.32NDD961 pKa = 4.93GILDD965 pKa = 3.53VDD967 pKa = 4.03EE968 pKa = 5.67GLVCTGANQGEE979 pKa = 4.37FKK981 pKa = 10.32IGYY984 pKa = 9.58VDD986 pKa = 3.43TTVGKK991 pKa = 8.43TGLMEE996 pKa = 4.78NMLTNTDD1003 pKa = 3.15NFGPDD1008 pKa = 3.1GVYY1011 pKa = 10.96DD1012 pKa = 4.6KK1013 pKa = 10.84IPNVTLVPYY1022 pKa = 10.41AINDD1026 pKa = 3.35VSEE1029 pKa = 4.08TRR1031 pKa = 11.84LIEE1034 pKa = 4.1DD1035 pKa = 3.82QIDD1038 pKa = 3.61VFYY1041 pKa = 10.85LGSSTEE1047 pKa = 4.17GDD1049 pKa = 3.91FLNQTGSNSQKK1060 pKa = 10.63LPSTLNSTVRR1070 pKa = 11.84AWGEE1074 pKa = 4.01NNNKK1078 pKa = 9.84SVLVVQNNAIDD1089 pKa = 3.76FGYY1092 pKa = 10.72RR1093 pKa = 11.84LVNNSVNPNTPYY1105 pKa = 10.8DD1106 pKa = 3.72ALGEE1110 pKa = 4.31SVMTNGYY1117 pKa = 8.94WPVSTFDD1124 pKa = 3.27QTGSVQLTASSATEE1138 pKa = 4.01RR1139 pKa = 11.84YY1140 pKa = 9.76DD1141 pKa = 4.73VVMTDD1146 pKa = 3.52KK1147 pKa = 11.15NGKK1150 pKa = 8.35ATLLSSRR1157 pKa = 11.84TSNMVLLPDD1166 pKa = 3.57ATIMKK1171 pKa = 10.25DD1172 pKa = 2.89NSTQSTVSTDD1182 pKa = 2.84ILKK1185 pKa = 10.5VVANIHH1191 pKa = 6.74AYY1193 pKa = 10.41LFDD1196 pKa = 4.06MFLATPQCKK1205 pKa = 10.47GIDD1208 pKa = 3.57TDD1210 pKa = 4.12GDD1212 pKa = 3.87GVANHH1217 pKa = 7.28LDD1219 pKa = 3.86LDD1221 pKa = 4.23SDD1223 pKa = 4.4GDD1225 pKa = 3.87GCPDD1229 pKa = 3.09IKK1231 pKa = 10.86EE1232 pKa = 4.29AGISNYY1238 pKa = 9.7LFKK1241 pKa = 10.92NDD1243 pKa = 3.75QEE1245 pKa = 4.71SLMVSGEE1252 pKa = 4.19VVNLSEE1258 pKa = 4.73TDD1260 pKa = 3.25STQNTTTTIGMSRR1273 pKa = 11.84MNPSNGGDD1281 pKa = 3.04NVGPFNGFHH1290 pKa = 7.78DD1291 pKa = 4.61YY1292 pKa = 11.41LEE1294 pKa = 4.19QTSPGVYY1301 pKa = 10.0EE1302 pKa = 3.86NAPYY1306 pKa = 10.27QLEE1309 pKa = 4.62PYY1311 pKa = 10.4LSNEE1315 pKa = 3.78VNACRR1320 pKa = 11.84IRR1322 pKa = 11.84KK1323 pKa = 9.26IYY1325 pKa = 9.2TNPAMHH1331 pKa = 7.23TPRR1334 pKa = 11.84KK1335 pKa = 9.1AQQ1337 pKa = 3.08
MM1 pKa = 7.48HH2 pKa = 7.69KK3 pKa = 10.05ISYY6 pKa = 10.1ISILVVCAILVGYY19 pKa = 8.41VPKK22 pKa = 10.79DD23 pKa = 3.47VVSNSGVKK31 pKa = 10.1VEE33 pKa = 4.52SVRR36 pKa = 11.84KK37 pKa = 9.85NINSDD42 pKa = 2.66TDD44 pKa = 3.62YY45 pKa = 11.86DD46 pKa = 4.05EE47 pKa = 6.96DD48 pKa = 5.34GVLNDD53 pKa = 5.25NDD55 pKa = 5.49LDD57 pKa = 4.43DD58 pKa = 6.26DD59 pKa = 4.41NDD61 pKa = 4.89GILDD65 pKa = 3.44IDD67 pKa = 3.85EE68 pKa = 5.61GFVCTTNSQKK78 pKa = 9.51TVLKK82 pKa = 10.39AISNNIPAEE91 pKa = 4.18GTSYY95 pKa = 11.14CGGLKK100 pKa = 10.28NSYY103 pKa = 7.4THH105 pKa = 7.2FSDD108 pKa = 6.02GYY110 pKa = 11.1LDD112 pKa = 3.73TQNDD116 pKa = 3.07ITGTYY121 pKa = 7.87TLNYY125 pKa = 9.1SYY127 pKa = 11.37SGLDD131 pKa = 3.29VSKK134 pKa = 10.75EE135 pKa = 3.87AIFQHH140 pKa = 6.17RR141 pKa = 11.84LVYY144 pKa = 10.83SNVNGSHH151 pKa = 6.5GVDD154 pKa = 2.97QNTSNVRR161 pKa = 11.84ITSKK165 pKa = 10.35IYY167 pKa = 10.73VNNTLVRR174 pKa = 11.84TASVTGWNYY183 pKa = 10.09HH184 pKa = 4.5SQPRR188 pKa = 11.84EE189 pKa = 3.67KK190 pKa = 10.7DD191 pKa = 2.89NKK193 pKa = 10.02YY194 pKa = 11.09YY195 pKa = 11.05YY196 pKa = 7.28FTPSTASGTVRR207 pKa = 11.84IDD209 pKa = 3.06ITFEE213 pKa = 3.96RR214 pKa = 11.84LNGYY218 pKa = 9.89CDD220 pKa = 3.98VIPEE224 pKa = 4.18VFFGGNLSQTSDD236 pKa = 4.79PICTSIDD243 pKa = 3.37TDD245 pKa = 3.69GDD247 pKa = 4.4GIPDD251 pKa = 3.97HH252 pKa = 7.11QDD254 pKa = 2.93LDD256 pKa = 4.2SDD258 pKa = 4.88GDD260 pKa = 3.95GCPDD264 pKa = 3.36ALEE267 pKa = 4.51GQGGFTAADD276 pKa = 3.81LVSSALAGGNSGANYY291 pKa = 8.97TGVAGSVVSNLGATVGSNGVPTKK314 pKa = 10.41AASGQGRR321 pKa = 11.84GTSRR325 pKa = 11.84EE326 pKa = 3.96IQQVASACIGCSTLAGDD343 pKa = 3.97VTDD346 pKa = 3.92TGGDD350 pKa = 3.53PVGNAMIRR358 pKa = 11.84VYY360 pKa = 11.41ADD362 pKa = 3.61FDD364 pKa = 4.14QNGNISSGDD373 pKa = 3.52EE374 pKa = 3.75LLKK377 pKa = 10.93VVNADD382 pKa = 3.09AQGAYY387 pKa = 10.14SINKK391 pKa = 9.24GYY393 pKa = 10.69YY394 pKa = 8.11ITVKK398 pKa = 10.67DD399 pKa = 4.45DD400 pKa = 3.65FSGDD404 pKa = 3.47VFSGNNNSSGDD415 pKa = 3.59YY416 pKa = 10.27NAWLDD421 pKa = 4.39ASWTQSNSANSGISSNTNGDD441 pKa = 3.73DD442 pKa = 3.73NLEE445 pKa = 3.98SRR447 pKa = 11.84HH448 pKa = 6.6IYY450 pKa = 8.77IHH452 pKa = 6.74NNTQVYY458 pKa = 9.58RR459 pKa = 11.84AVNLSKK465 pKa = 10.91FKK467 pKa = 9.86TANLRR472 pKa = 11.84FDD474 pKa = 4.25YY475 pKa = 9.39DD476 pKa = 3.77TEE478 pKa = 4.59NDD480 pKa = 3.58LDD482 pKa = 3.93ATDD485 pKa = 4.0KK486 pKa = 11.19FHH488 pKa = 8.01VEE490 pKa = 3.84LSKK493 pKa = 11.44DD494 pKa = 3.12GGATYY499 pKa = 7.68TTVYY503 pKa = 9.93TYY505 pKa = 11.24TDD507 pKa = 3.81GNVDD511 pKa = 3.54NQPARR516 pKa = 11.84SQNINLNGYY525 pKa = 9.3AGEE528 pKa = 4.29SNVIIRR534 pKa = 11.84FRR536 pKa = 11.84AEE538 pKa = 3.96SSSGEE543 pKa = 4.37YY544 pKa = 9.61IWLDD548 pKa = 3.01NVTIYY553 pKa = 11.15NAFPTVLLKK562 pKa = 10.24PVYY565 pKa = 9.43NEE567 pKa = 4.58PYY569 pKa = 9.83RR570 pKa = 11.84SSNPATIAVNPSTTTCSGLDD590 pKa = 3.52FTLLDD595 pKa = 4.26ACDD598 pKa = 3.99VDD600 pKa = 4.16PTDD603 pKa = 3.32TDD605 pKa = 3.72GDD607 pKa = 4.72GVCDD611 pKa = 6.08AIDD614 pKa = 4.72IDD616 pKa = 5.27DD617 pKa = 5.49DD618 pKa = 4.87DD619 pKa = 7.17DD620 pKa = 4.5GVLDD624 pKa = 3.58EE625 pKa = 5.13TEE627 pKa = 4.05GHH629 pKa = 6.4FCGEE633 pKa = 4.27LDD635 pKa = 3.51RR636 pKa = 11.84NIIIGYY642 pKa = 9.19LDD644 pKa = 3.55SGVGNDD650 pKa = 4.51GLATDD655 pKa = 4.21MLLNLKK661 pKa = 10.4NYY663 pKa = 10.18GEE665 pKa = 4.45YY666 pKa = 9.54GTYY669 pKa = 10.54DD670 pKa = 3.13KK671 pKa = 11.44VRR673 pKa = 11.84GVVLVPFSSSAEE685 pKa = 3.67ITEE688 pKa = 4.71ANLEE692 pKa = 4.17ANNIDD697 pKa = 3.65VFFAGSSTANSEE709 pKa = 3.98GSSDD713 pKa = 3.79KK714 pKa = 11.12LSSSTNAVLMSWAEE728 pKa = 4.04NNDD731 pKa = 2.96KK732 pKa = 11.27GLFVLQNNAVDD743 pKa = 3.55YY744 pKa = 10.85GYY746 pKa = 8.48TLTNNDD752 pKa = 4.8LPPDD756 pKa = 3.83VPNGEE761 pKa = 4.71LGYY764 pKa = 10.22QVYY767 pKa = 9.63TNGYY771 pKa = 8.24WPVSSITQTGTVKK784 pKa = 8.23MTISSSTRR792 pKa = 11.84SFEE795 pKa = 4.27ALMVDD800 pKa = 3.84SKK802 pKa = 11.61SRR804 pKa = 11.84PTLVKK809 pKa = 10.73DD810 pKa = 3.64NMMKK814 pKa = 10.64LVIFPDD820 pKa = 3.13ATIYY824 pKa = 10.87NDD826 pKa = 3.38EE827 pKa = 4.89ANTVDD832 pKa = 4.21PNKK835 pKa = 10.66NDD837 pKa = 3.37ATKK840 pKa = 10.84VIADD844 pKa = 3.79TWAFVMDD851 pKa = 3.88TFIGDD856 pKa = 3.72GCSEE860 pKa = 5.02LDD862 pKa = 3.67TNGDD866 pKa = 3.54GTPNHH871 pKa = 7.18LDD873 pKa = 3.69LDD875 pKa = 4.25SDD877 pKa = 4.55GDD879 pKa = 3.99GCSDD883 pKa = 3.48AFEE886 pKa = 5.01ANATEE891 pKa = 5.7DD892 pKa = 3.3ITPNFQFDD900 pKa = 3.97VQKK903 pKa = 10.22GTSSDD908 pKa = 3.62TNGDD912 pKa = 3.62GLADD916 pKa = 3.93AVDD919 pKa = 3.97EE920 pKa = 4.43NLNGIPDD927 pKa = 4.14YY928 pKa = 11.42VSAYY932 pKa = 10.65YY933 pKa = 9.89PDD935 pKa = 5.11ALDD938 pKa = 3.75NTTCTDD944 pKa = 3.12SDD946 pKa = 3.74GDD948 pKa = 4.34TIVDD952 pKa = 4.73AVDD955 pKa = 5.13LDD957 pKa = 4.27DD958 pKa = 6.61DD959 pKa = 4.32NDD961 pKa = 4.93GILDD965 pKa = 3.53VDD967 pKa = 4.03EE968 pKa = 5.67GLVCTGANQGEE979 pKa = 4.37FKK981 pKa = 10.32IGYY984 pKa = 9.58VDD986 pKa = 3.43TTVGKK991 pKa = 8.43TGLMEE996 pKa = 4.78NMLTNTDD1003 pKa = 3.15NFGPDD1008 pKa = 3.1GVYY1011 pKa = 10.96DD1012 pKa = 4.6KK1013 pKa = 10.84IPNVTLVPYY1022 pKa = 10.41AINDD1026 pKa = 3.35VSEE1029 pKa = 4.08TRR1031 pKa = 11.84LIEE1034 pKa = 4.1DD1035 pKa = 3.82QIDD1038 pKa = 3.61VFYY1041 pKa = 10.85LGSSTEE1047 pKa = 4.17GDD1049 pKa = 3.91FLNQTGSNSQKK1060 pKa = 10.63LPSTLNSTVRR1070 pKa = 11.84AWGEE1074 pKa = 4.01NNNKK1078 pKa = 9.84SVLVVQNNAIDD1089 pKa = 3.76FGYY1092 pKa = 10.72RR1093 pKa = 11.84LVNNSVNPNTPYY1105 pKa = 10.8DD1106 pKa = 3.72ALGEE1110 pKa = 4.31SVMTNGYY1117 pKa = 8.94WPVSTFDD1124 pKa = 3.27QTGSVQLTASSATEE1138 pKa = 4.01RR1139 pKa = 11.84YY1140 pKa = 9.76DD1141 pKa = 4.73VVMTDD1146 pKa = 3.52KK1147 pKa = 11.15NGKK1150 pKa = 8.35ATLLSSRR1157 pKa = 11.84TSNMVLLPDD1166 pKa = 3.57ATIMKK1171 pKa = 10.25DD1172 pKa = 2.89NSTQSTVSTDD1182 pKa = 2.84ILKK1185 pKa = 10.5VVANIHH1191 pKa = 6.74AYY1193 pKa = 10.41LFDD1196 pKa = 4.06MFLATPQCKK1205 pKa = 10.47GIDD1208 pKa = 3.57TDD1210 pKa = 4.12GDD1212 pKa = 3.87GVANHH1217 pKa = 7.28LDD1219 pKa = 3.86LDD1221 pKa = 4.23SDD1223 pKa = 4.4GDD1225 pKa = 3.87GCPDD1229 pKa = 3.09IKK1231 pKa = 10.86EE1232 pKa = 4.29AGISNYY1238 pKa = 9.7LFKK1241 pKa = 10.92NDD1243 pKa = 3.75QEE1245 pKa = 4.71SLMVSGEE1252 pKa = 4.19VVNLSEE1258 pKa = 4.73TDD1260 pKa = 3.25STQNTTTTIGMSRR1273 pKa = 11.84MNPSNGGDD1281 pKa = 3.04NVGPFNGFHH1290 pKa = 7.78DD1291 pKa = 4.61YY1292 pKa = 11.41LEE1294 pKa = 4.19QTSPGVYY1301 pKa = 10.0EE1302 pKa = 3.86NAPYY1306 pKa = 10.27QLEE1309 pKa = 4.62PYY1311 pKa = 10.4LSNEE1315 pKa = 3.78VNACRR1320 pKa = 11.84IRR1322 pKa = 11.84KK1323 pKa = 9.26IYY1325 pKa = 9.2TNPAMHH1331 pKa = 7.23TPRR1334 pKa = 11.84KK1335 pKa = 9.1AQQ1337 pKa = 3.08
Molecular weight: 143.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0FW70|L0FW70_ECHVK Small-conductance mechanosensitive channel OS=Echinicola vietnamensis (strain DSM 17526 / LMG 23754 / KMM 6221) OX=926556 GN=Echvi_1017 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.44RR21 pKa = 11.84MASPNGRR28 pKa = 11.84RR29 pKa = 11.84VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.48GRR39 pKa = 11.84HH40 pKa = 5.05KK41 pKa = 10.93LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.44RR21 pKa = 11.84MASPNGRR28 pKa = 11.84RR29 pKa = 11.84VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.48GRR39 pKa = 11.84HH40 pKa = 5.05KK41 pKa = 10.93LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1596854 |
29 |
6228 |
354.1 |
39.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.066 ± 0.034 | 0.72 ± 0.012 |
5.728 ± 0.029 | 6.852 ± 0.037 |
4.866 ± 0.028 | 7.246 ± 0.038 |
2.094 ± 0.021 | 6.633 ± 0.032 |
6.556 ± 0.05 | 9.429 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.567 ± 0.019 | 4.912 ± 0.033 |
3.969 ± 0.016 | 3.759 ± 0.02 |
4.125 ± 0.027 | 6.25 ± 0.027 |
5.359 ± 0.047 | 6.503 ± 0.031 |
1.303 ± 0.014 | 4.063 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
