
Gordonia phage Suerte
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Nymbaxtervirinae; unclassified Nymbaxtervirinae
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
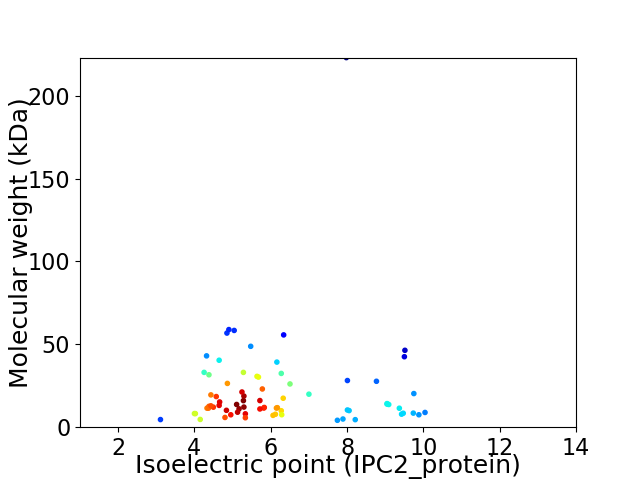
Virtual 2D-PAGE plot for 74 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5P8DDD4|A0A5P8DDD4_9CAUD Helix-turn-helix DNA binding protein OS=Gordonia phage Suerte OX=2652883 GN=35 PE=4 SV=1
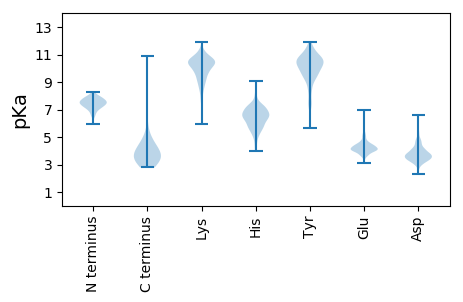
MM1 pKa = 7.11TAPEE5 pKa = 4.48RR6 pKa = 11.84MTPLASGVEE15 pKa = 4.07NGVRR19 pKa = 11.84WVTCVAPIYY28 pKa = 10.31GAVNGYY34 pKa = 9.27VLLPEE39 pKa = 3.98GHH41 pKa = 7.18PWRR44 pKa = 11.84GLDD47 pKa = 3.35YY48 pKa = 11.06DD49 pKa = 3.88QIDD52 pKa = 4.08VEE54 pKa = 4.55VHH56 pKa = 5.95GGLTYY61 pKa = 10.9GHH63 pKa = 7.31DD64 pKa = 3.15GWIGFDD70 pKa = 3.44TLHH73 pKa = 7.13AGDD76 pKa = 4.32VWPGSPQYY84 pKa = 10.73GGPRR88 pKa = 11.84PYY90 pKa = 11.42DD91 pKa = 3.48MRR93 pKa = 11.84WTAEE97 pKa = 3.92MVAEE101 pKa = 4.32EE102 pKa = 4.33ARR104 pKa = 11.84RR105 pKa = 11.84LAARR109 pKa = 11.84VAAAAEE115 pKa = 4.37TTDD118 pKa = 3.63TGPVSGPPYY127 pKa = 10.27PPDD130 pKa = 3.82RR131 pKa = 11.84VPDD134 pKa = 3.76DD135 pKa = 4.8HH136 pKa = 8.05DD137 pKa = 5.22DD138 pKa = 3.6GTMPTNVAEE147 pKa = 4.5FAEE150 pKa = 4.44QVVGRR155 pKa = 11.84RR156 pKa = 11.84IIKK159 pKa = 10.19AEE161 pKa = 3.85KK162 pKa = 9.13TDD164 pKa = 4.14FRR166 pKa = 11.84PEE168 pKa = 4.11DD169 pKa = 3.63FDD171 pKa = 4.21GTEE174 pKa = 3.61HH175 pKa = 6.72TYY177 pKa = 9.33WSSYY181 pKa = 10.55SDD183 pKa = 3.16THH185 pKa = 7.87GLVLTLDD192 pKa = 3.67DD193 pKa = 3.62GRR195 pKa = 11.84RR196 pKa = 11.84VVLVDD201 pKa = 3.84TSDD204 pKa = 3.51CCAYY208 pKa = 9.81TSLEE212 pKa = 4.01EE213 pKa = 4.45FFLHH217 pKa = 6.73PEE219 pKa = 4.11RR220 pKa = 11.84VDD222 pKa = 3.54HH223 pKa = 6.71VITGVATTDD232 pKa = 3.57GYY234 pKa = 10.89EE235 pKa = 3.94RR236 pKa = 11.84WHH238 pKa = 6.21VFADD242 pKa = 3.42LGDD245 pKa = 3.76ILEE248 pKa = 4.77LKK250 pKa = 10.3IGWSAGNPFYY260 pKa = 10.93YY261 pKa = 10.41GYY263 pKa = 10.5GFNIGVSNVIEE274 pKa = 4.5GSLAQPEE281 pKa = 4.53LEE283 pKa = 4.24QGQSS287 pKa = 2.93
MM1 pKa = 7.11TAPEE5 pKa = 4.48RR6 pKa = 11.84MTPLASGVEE15 pKa = 4.07NGVRR19 pKa = 11.84WVTCVAPIYY28 pKa = 10.31GAVNGYY34 pKa = 9.27VLLPEE39 pKa = 3.98GHH41 pKa = 7.18PWRR44 pKa = 11.84GLDD47 pKa = 3.35YY48 pKa = 11.06DD49 pKa = 3.88QIDD52 pKa = 4.08VEE54 pKa = 4.55VHH56 pKa = 5.95GGLTYY61 pKa = 10.9GHH63 pKa = 7.31DD64 pKa = 3.15GWIGFDD70 pKa = 3.44TLHH73 pKa = 7.13AGDD76 pKa = 4.32VWPGSPQYY84 pKa = 10.73GGPRR88 pKa = 11.84PYY90 pKa = 11.42DD91 pKa = 3.48MRR93 pKa = 11.84WTAEE97 pKa = 3.92MVAEE101 pKa = 4.32EE102 pKa = 4.33ARR104 pKa = 11.84RR105 pKa = 11.84LAARR109 pKa = 11.84VAAAAEE115 pKa = 4.37TTDD118 pKa = 3.63TGPVSGPPYY127 pKa = 10.27PPDD130 pKa = 3.82RR131 pKa = 11.84VPDD134 pKa = 3.76DD135 pKa = 4.8HH136 pKa = 8.05DD137 pKa = 5.22DD138 pKa = 3.6GTMPTNVAEE147 pKa = 4.5FAEE150 pKa = 4.44QVVGRR155 pKa = 11.84RR156 pKa = 11.84IIKK159 pKa = 10.19AEE161 pKa = 3.85KK162 pKa = 9.13TDD164 pKa = 4.14FRR166 pKa = 11.84PEE168 pKa = 4.11DD169 pKa = 3.63FDD171 pKa = 4.21GTEE174 pKa = 3.61HH175 pKa = 6.72TYY177 pKa = 9.33WSSYY181 pKa = 10.55SDD183 pKa = 3.16THH185 pKa = 7.87GLVLTLDD192 pKa = 3.67DD193 pKa = 3.62GRR195 pKa = 11.84RR196 pKa = 11.84VVLVDD201 pKa = 3.84TSDD204 pKa = 3.51CCAYY208 pKa = 9.81TSLEE212 pKa = 4.01EE213 pKa = 4.45FFLHH217 pKa = 6.73PEE219 pKa = 4.11RR220 pKa = 11.84VDD222 pKa = 3.54HH223 pKa = 6.71VITGVATTDD232 pKa = 3.57GYY234 pKa = 10.89EE235 pKa = 3.94RR236 pKa = 11.84WHH238 pKa = 6.21VFADD242 pKa = 3.42LGDD245 pKa = 3.76ILEE248 pKa = 4.77LKK250 pKa = 10.3IGWSAGNPFYY260 pKa = 10.93YY261 pKa = 10.41GYY263 pKa = 10.5GFNIGVSNVIEE274 pKa = 4.5GSLAQPEE281 pKa = 4.53LEE283 pKa = 4.24QGQSS287 pKa = 2.93
Molecular weight: 31.62 kDa
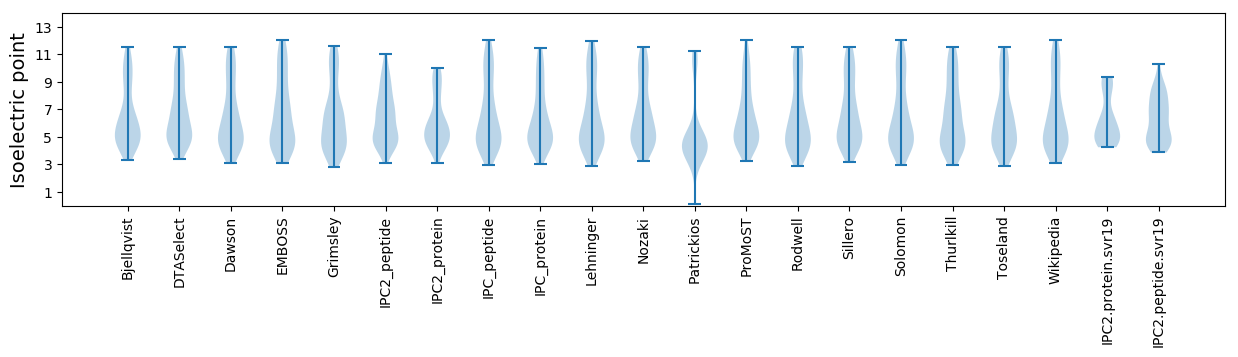
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5P8DDH2|A0A5P8DDH2_9CAUD Tail assembly chaperone OS=Gordonia phage Suerte OX=2652883 GN=15 PE=4 SV=1
MM1 pKa = 7.59ARR3 pKa = 11.84PPLPLGTWGKK13 pKa = 8.09ITRR16 pKa = 11.84KK17 pKa = 7.83EE18 pKa = 4.03TAPGRR23 pKa = 11.84WRR25 pKa = 11.84ARR27 pKa = 11.84AQYY30 pKa = 10.63RR31 pKa = 11.84DD32 pKa = 3.45FDD34 pKa = 4.04GVTRR38 pKa = 11.84PVEE41 pKa = 4.1RR42 pKa = 11.84YY43 pKa = 8.93GKK45 pKa = 10.11SGAAAEE51 pKa = 4.2RR52 pKa = 11.84ALVEE56 pKa = 4.04ALRR59 pKa = 11.84DD60 pKa = 3.35RR61 pKa = 11.84AAARR65 pKa = 11.84GDD67 pKa = 3.48IINRR71 pKa = 11.84DD72 pKa = 3.34TTIAALATAWIASRR86 pKa = 11.84VAEE89 pKa = 4.3DD90 pKa = 3.47ALRR93 pKa = 11.84EE94 pKa = 3.98QSVEE98 pKa = 4.09QYY100 pKa = 10.59QSAIDD105 pKa = 3.64KK106 pKa = 10.6HH107 pKa = 6.23IVPALGRR114 pKa = 11.84VRR116 pKa = 11.84IGEE119 pKa = 4.26VGVGILDD126 pKa = 3.71RR127 pKa = 11.84FLEE130 pKa = 5.37GISSAAVAKK139 pKa = 9.94RR140 pKa = 11.84CRR142 pKa = 11.84VVLTGMFGLAARR154 pKa = 11.84HH155 pKa = 6.19DD156 pKa = 5.3AIDD159 pKa = 3.87HH160 pKa = 5.53NPVRR164 pKa = 11.84EE165 pKa = 4.24TVSRR169 pKa = 11.84AQKK172 pKa = 9.78RR173 pKa = 11.84IPVRR177 pKa = 11.84AMTVGEE183 pKa = 4.3VAVMRR188 pKa = 11.84ARR190 pKa = 11.84VAAWAGANASGPPRR204 pKa = 11.84GQDD207 pKa = 3.65LPDD210 pKa = 4.39ILDD213 pKa = 3.64AMLGTGARR221 pKa = 11.84IGEE224 pKa = 3.98VLAIRR229 pKa = 11.84KK230 pKa = 8.91EE231 pKa = 3.99DD232 pKa = 3.52LDD234 pKa = 4.2LRR236 pKa = 11.84SNPPTVVITGTIVGNKK252 pKa = 8.52RR253 pKa = 11.84QPAPKK258 pKa = 8.67TDD260 pKa = 3.19YY261 pKa = 10.73SYY263 pKa = 11.55RR264 pKa = 11.84RR265 pKa = 11.84LILPSFVAAALRR277 pKa = 11.84RR278 pKa = 11.84QLARR282 pKa = 11.84NLPTDD287 pKa = 4.05DD288 pKa = 3.8LHH290 pKa = 9.05LVFPSRR296 pKa = 11.84TGGPRR301 pKa = 11.84MGNNVRR307 pKa = 11.84RR308 pKa = 11.84QLRR311 pKa = 11.84EE312 pKa = 3.62ARR314 pKa = 11.84GDD316 pKa = 3.57DD317 pKa = 4.09FDD319 pKa = 3.91WVTPKK324 pKa = 10.51VYY326 pKa = 10.5RR327 pKa = 11.84KK328 pKa = 7.56TVATAIDD335 pKa = 3.53RR336 pKa = 11.84AADD339 pKa = 3.11IEE341 pKa = 4.41TAARR345 pKa = 11.84QLGHH349 pKa = 6.24SRR351 pKa = 11.84STTTQAHH358 pKa = 4.56YY359 pKa = 10.73VEE361 pKa = 4.99RR362 pKa = 11.84LLVAPDD368 pKa = 3.29VRR370 pKa = 11.84DD371 pKa = 3.52VLDD374 pKa = 3.44EE375 pKa = 4.46FGPVSRR381 pKa = 11.84GFSVGDD387 pKa = 3.33VSS389 pKa = 4.72
MM1 pKa = 7.59ARR3 pKa = 11.84PPLPLGTWGKK13 pKa = 8.09ITRR16 pKa = 11.84KK17 pKa = 7.83EE18 pKa = 4.03TAPGRR23 pKa = 11.84WRR25 pKa = 11.84ARR27 pKa = 11.84AQYY30 pKa = 10.63RR31 pKa = 11.84DD32 pKa = 3.45FDD34 pKa = 4.04GVTRR38 pKa = 11.84PVEE41 pKa = 4.1RR42 pKa = 11.84YY43 pKa = 8.93GKK45 pKa = 10.11SGAAAEE51 pKa = 4.2RR52 pKa = 11.84ALVEE56 pKa = 4.04ALRR59 pKa = 11.84DD60 pKa = 3.35RR61 pKa = 11.84AAARR65 pKa = 11.84GDD67 pKa = 3.48IINRR71 pKa = 11.84DD72 pKa = 3.34TTIAALATAWIASRR86 pKa = 11.84VAEE89 pKa = 4.3DD90 pKa = 3.47ALRR93 pKa = 11.84EE94 pKa = 3.98QSVEE98 pKa = 4.09QYY100 pKa = 10.59QSAIDD105 pKa = 3.64KK106 pKa = 10.6HH107 pKa = 6.23IVPALGRR114 pKa = 11.84VRR116 pKa = 11.84IGEE119 pKa = 4.26VGVGILDD126 pKa = 3.71RR127 pKa = 11.84FLEE130 pKa = 5.37GISSAAVAKK139 pKa = 9.94RR140 pKa = 11.84CRR142 pKa = 11.84VVLTGMFGLAARR154 pKa = 11.84HH155 pKa = 6.19DD156 pKa = 5.3AIDD159 pKa = 3.87HH160 pKa = 5.53NPVRR164 pKa = 11.84EE165 pKa = 4.24TVSRR169 pKa = 11.84AQKK172 pKa = 9.78RR173 pKa = 11.84IPVRR177 pKa = 11.84AMTVGEE183 pKa = 4.3VAVMRR188 pKa = 11.84ARR190 pKa = 11.84VAAWAGANASGPPRR204 pKa = 11.84GQDD207 pKa = 3.65LPDD210 pKa = 4.39ILDD213 pKa = 3.64AMLGTGARR221 pKa = 11.84IGEE224 pKa = 3.98VLAIRR229 pKa = 11.84KK230 pKa = 8.91EE231 pKa = 3.99DD232 pKa = 3.52LDD234 pKa = 4.2LRR236 pKa = 11.84SNPPTVVITGTIVGNKK252 pKa = 8.52RR253 pKa = 11.84QPAPKK258 pKa = 8.67TDD260 pKa = 3.19YY261 pKa = 10.73SYY263 pKa = 11.55RR264 pKa = 11.84RR265 pKa = 11.84LILPSFVAAALRR277 pKa = 11.84RR278 pKa = 11.84QLARR282 pKa = 11.84NLPTDD287 pKa = 4.05DD288 pKa = 3.8LHH290 pKa = 9.05LVFPSRR296 pKa = 11.84TGGPRR301 pKa = 11.84MGNNVRR307 pKa = 11.84RR308 pKa = 11.84QLRR311 pKa = 11.84EE312 pKa = 3.62ARR314 pKa = 11.84GDD316 pKa = 3.57DD317 pKa = 4.09FDD319 pKa = 3.91WVTPKK324 pKa = 10.51VYY326 pKa = 10.5RR327 pKa = 11.84KK328 pKa = 7.56TVATAIDD335 pKa = 3.53RR336 pKa = 11.84AADD339 pKa = 3.11IEE341 pKa = 4.41TAARR345 pKa = 11.84QLGHH349 pKa = 6.24SRR351 pKa = 11.84STTTQAHH358 pKa = 4.56YY359 pKa = 10.73VEE361 pKa = 4.99RR362 pKa = 11.84LLVAPDD368 pKa = 3.29VRR370 pKa = 11.84DD371 pKa = 3.52VLDD374 pKa = 3.44EE375 pKa = 4.46FGPVSRR381 pKa = 11.84GFSVGDD387 pKa = 3.33VSS389 pKa = 4.72
Molecular weight: 42.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14950 |
38 |
2137 |
202.0 |
21.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.575 ± 0.548 | 0.696 ± 0.124 |
6.749 ± 0.407 | 5.579 ± 0.252 |
2.408 ± 0.162 | 8.415 ± 0.46 |
1.96 ± 0.2 | 4.589 ± 0.204 |
3.157 ± 0.255 | 7.819 ± 0.227 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.047 ± 0.124 | 3.043 ± 0.24 |
5.679 ± 0.284 | 3.726 ± 0.328 |
7.505 ± 0.467 | 5.344 ± 0.229 |
7.344 ± 0.27 | 7.324 ± 0.293 |
1.853 ± 0.144 | 2.187 ± 0.135 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
