
Eptesicus fuscus gammaherpesvirus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; unclassified Gammaherpesvirinae
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
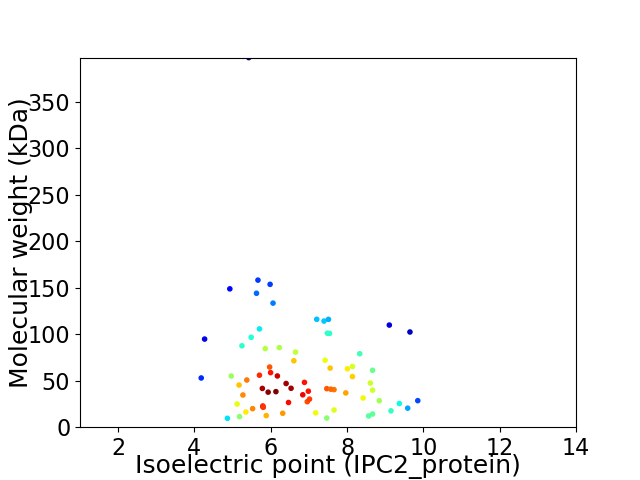
Virtual 2D-PAGE plot for 75 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
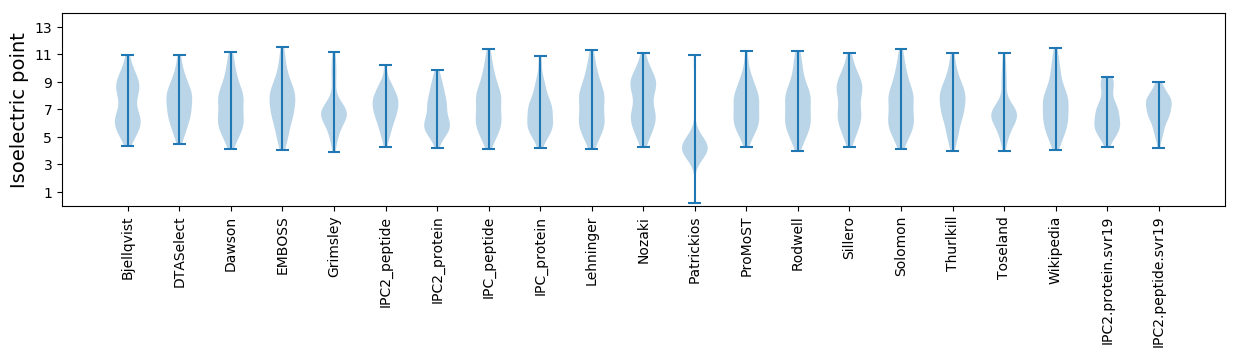
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D0ZNY5|A0A2D0ZNY5_9GAMA Uncharacterized protein OS=Eptesicus fuscus gammaherpesvirus OX=2035399 PE=4 SV=1
MM1 pKa = 7.03ATAVPIWGVSEE12 pKa = 4.43ADD14 pKa = 3.29GAEE17 pKa = 3.97WNNLIEE23 pKa = 4.52LGVRR27 pKa = 11.84NGSQMEE33 pKa = 4.11RR34 pKa = 11.84AVSILRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84LYY44 pKa = 8.9NTEE47 pKa = 3.73PLGYY51 pKa = 9.78LASLLALRR59 pKa = 11.84QEE61 pKa = 4.59LNSVTGGSAQEE72 pKa = 4.01ILLEE76 pKa = 4.11SLAVIQQLAKK86 pKa = 10.56VIYY89 pKa = 10.42GEE91 pKa = 5.68LKK93 pKa = 10.45DD94 pKa = 3.87QPPTARR100 pKa = 11.84INEE103 pKa = 4.01IFRR106 pKa = 11.84DD107 pKa = 3.73CRR109 pKa = 11.84LRR111 pKa = 11.84LALVLDD117 pKa = 4.71AACGCVDD124 pKa = 4.33CFRR127 pKa = 11.84LAKK130 pKa = 9.37VLKK133 pKa = 9.67VAPRR137 pKa = 11.84CLRR140 pKa = 11.84PPKK143 pKa = 10.56LMAHH147 pKa = 6.5EE148 pKa = 4.42RR149 pKa = 11.84QCAAASTLTYY159 pKa = 9.8FYY161 pKa = 10.66NSYY164 pKa = 9.43MVAGSDD170 pKa = 3.64VEE172 pKa = 4.3VAPEE176 pKa = 4.44HH177 pKa = 7.13IEE179 pKa = 3.98LLLIRR184 pKa = 11.84PDD186 pKa = 3.53QLNSYY191 pKa = 9.32PYY193 pKa = 10.38GWDD196 pKa = 3.35EE197 pKa = 4.04EE198 pKa = 4.9VAGLMSCLNLCWLYY212 pKa = 11.12TMMYY216 pKa = 8.73EE217 pKa = 3.84HH218 pKa = 7.57ATRR221 pKa = 11.84GMCTLTRR228 pKa = 11.84ALEE231 pKa = 4.07EE232 pKa = 4.22AYY234 pKa = 10.52HH235 pKa = 6.03RR236 pKa = 11.84VRR238 pKa = 11.84PGSKK242 pKa = 8.73ATLEE246 pKa = 4.07NMVQAVIEE254 pKa = 4.4GDD256 pKa = 3.49SRR258 pKa = 11.84ASRR261 pKa = 11.84SEE263 pKa = 3.75EE264 pKa = 3.97PRR266 pKa = 11.84PHH268 pKa = 7.22HH269 pKa = 5.55YY270 pKa = 9.61TLIKK274 pKa = 10.15PLVTMKK280 pKa = 10.62HH281 pKa = 3.82EE282 pKa = 4.51HH283 pKa = 6.13RR284 pKa = 11.84WSEE287 pKa = 4.12KK288 pKa = 9.53QDD290 pKa = 3.43SAVQTQVSCLYY301 pKa = 9.88EE302 pKa = 4.2LVKK305 pKa = 10.84GMYY308 pKa = 9.89GPYY311 pKa = 10.1ARR313 pKa = 11.84QVQYY317 pKa = 11.09GRR319 pKa = 11.84KK320 pKa = 6.81PTDD323 pKa = 3.28MVPPLLSSVLMGVHH337 pKa = 7.02PEE339 pKa = 3.93LLRR342 pKa = 11.84EE343 pKa = 4.18KK344 pKa = 10.42DD345 pKa = 3.61PPLCQPLQPPLGSSSATGDD364 pKa = 3.23KK365 pKa = 11.1AGDD368 pKa = 3.55VSAEE372 pKa = 3.9RR373 pKa = 11.84AAGRR377 pKa = 11.84ASEE380 pKa = 4.53AEE382 pKa = 4.01GDD384 pKa = 3.78RR385 pKa = 11.84QYY387 pKa = 8.06TTCGCLGACACGTLDD402 pKa = 3.46TTEE405 pKa = 4.28GEE407 pKa = 4.54IVYY410 pKa = 9.79HH411 pKa = 6.51AARR414 pKa = 11.84GTGPKK419 pKa = 9.48PFCLGTGSGQIPCRR433 pKa = 11.84GEE435 pKa = 3.94DD436 pKa = 3.29VRR438 pKa = 11.84DD439 pKa = 3.77ARR441 pKa = 11.84DD442 pKa = 3.53PTCQRR447 pKa = 11.84EE448 pKa = 4.56AIGEE452 pKa = 4.28VTEE455 pKa = 5.53GADD458 pKa = 3.97QVDD461 pKa = 3.75KK462 pKa = 8.99TTTQRR467 pKa = 11.84AASQFEE473 pKa = 4.27PTPEE477 pKa = 3.66DD478 pKa = 3.82VIRR481 pKa = 11.84LGARR485 pKa = 11.84RR486 pKa = 11.84KK487 pKa = 8.11TSGIGTAAGDD497 pKa = 3.68VLRR500 pKa = 11.84PDD502 pKa = 3.55IAQNQYY508 pKa = 10.69PLLALDD514 pKa = 4.25PRR516 pKa = 11.84DD517 pKa = 3.6LEE519 pKa = 4.3SRR521 pKa = 11.84RR522 pKa = 11.84VQHH525 pKa = 5.78VFSDD529 pKa = 3.72EE530 pKa = 4.16RR531 pKa = 11.84AGSPEE536 pKa = 3.89TGEE539 pKa = 4.28PDD541 pKa = 3.11QEE543 pKa = 3.93PRR545 pKa = 11.84KK546 pKa = 10.01RR547 pKa = 11.84KK548 pKa = 9.25PEE550 pKa = 4.2SGNGSPGASPFFKK563 pKa = 10.56SIWGGGDD570 pKa = 3.39YY571 pKa = 10.78VRR573 pKa = 11.84QSPPLGYY580 pKa = 10.82GPTKK584 pKa = 10.63DD585 pKa = 3.08RR586 pKa = 11.84AVGDD590 pKa = 3.97TYY592 pKa = 11.26WNYY595 pKa = 10.74TGTDD599 pKa = 3.72DD600 pKa = 4.19EE601 pKa = 4.76QEE603 pKa = 3.91RR604 pKa = 11.84CEE606 pKa = 4.05ADD608 pKa = 3.37GVGDD612 pKa = 3.52EE613 pKa = 6.04DD614 pKa = 4.73EE615 pKa = 5.97YY616 pKa = 11.5EE617 pKa = 4.64DD618 pKa = 5.86CDD620 pKa = 4.38EE621 pKa = 5.62LDD623 pKa = 4.61LRR625 pKa = 11.84DD626 pKa = 3.88TKK628 pKa = 10.87TGGCHH633 pKa = 5.3ATLGGDD639 pKa = 4.18GNQEE643 pKa = 3.92PPFLLLRR650 pKa = 11.84DD651 pKa = 4.56FEE653 pKa = 5.37ALLKK657 pKa = 9.79TGSSEE662 pKa = 3.82GLSPHH667 pKa = 6.04IQVSHH672 pKa = 5.23GTEE675 pKa = 3.97KK676 pKa = 10.09EE677 pKa = 4.17TLEE680 pKa = 5.59DD681 pKa = 3.88GGSCLKK687 pKa = 10.87VYY689 pKa = 10.71EE690 pKa = 5.01DD691 pKa = 4.25DD692 pKa = 4.48SKK694 pKa = 11.86GEE696 pKa = 4.1SGGNPRR702 pKa = 11.84DD703 pKa = 3.46VTGNVEE709 pKa = 4.27DD710 pKa = 5.35AVDD713 pKa = 5.0GYY715 pKa = 11.8NSDD718 pKa = 4.6DD719 pKa = 5.12LGISCCQDD727 pKa = 2.37GWSGDD732 pKa = 3.61DD733 pKa = 4.01GEE735 pKa = 5.58AVDD738 pKa = 5.59TEE740 pKa = 5.49DD741 pKa = 3.99EE742 pKa = 4.09DD743 pKa = 4.79TYY745 pKa = 11.51TNQEE749 pKa = 3.83GCGGGVEE756 pKa = 4.2EE757 pKa = 5.97GEE759 pKa = 3.96DD760 pKa = 3.25HH761 pKa = 7.78GIGEE765 pKa = 4.48EE766 pKa = 3.99EE767 pKa = 4.7DD768 pKa = 4.95GGDD771 pKa = 3.19EE772 pKa = 4.38CYY774 pKa = 10.51EE775 pKa = 4.23DD776 pKa = 4.66EE777 pKa = 4.8GAQSSYY783 pKa = 10.07YY784 pKa = 10.78DD785 pKa = 3.62RR786 pKa = 11.84VAGDD790 pKa = 3.4NADD793 pKa = 3.73EE794 pKa = 4.68LGEE797 pKa = 4.0EE798 pKa = 4.23LGEE801 pKa = 4.35DD802 pKa = 3.84DD803 pKa = 5.41EE804 pKa = 6.09LGEE807 pKa = 4.08EE808 pKa = 4.4LGDD811 pKa = 3.74EE812 pKa = 4.25EE813 pKa = 5.23SVEE816 pKa = 5.1DD817 pKa = 4.52EE818 pKa = 4.63DD819 pKa = 5.99SGDD822 pKa = 3.73GGDD825 pKa = 3.98GEE827 pKa = 4.98EE828 pKa = 4.02VCEE831 pKa = 4.98DD832 pKa = 4.06YY833 pKa = 10.9EE834 pKa = 4.52VCDD837 pKa = 3.78GDD839 pKa = 4.3EE840 pKa = 4.12EE841 pKa = 5.04CEE843 pKa = 4.12EE844 pKa = 4.52GNDD847 pKa = 3.57TGLEE851 pKa = 4.05TLEE854 pKa = 5.18DD855 pKa = 4.08SVDD858 pKa = 3.68EE859 pKa = 4.5EE860 pKa = 4.29NMGIYY865 pKa = 9.98FRR867 pKa = 11.84EE868 pKa = 4.59HH869 pKa = 5.35II870 pKa = 4.38
MM1 pKa = 7.03ATAVPIWGVSEE12 pKa = 4.43ADD14 pKa = 3.29GAEE17 pKa = 3.97WNNLIEE23 pKa = 4.52LGVRR27 pKa = 11.84NGSQMEE33 pKa = 4.11RR34 pKa = 11.84AVSILRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84LYY44 pKa = 8.9NTEE47 pKa = 3.73PLGYY51 pKa = 9.78LASLLALRR59 pKa = 11.84QEE61 pKa = 4.59LNSVTGGSAQEE72 pKa = 4.01ILLEE76 pKa = 4.11SLAVIQQLAKK86 pKa = 10.56VIYY89 pKa = 10.42GEE91 pKa = 5.68LKK93 pKa = 10.45DD94 pKa = 3.87QPPTARR100 pKa = 11.84INEE103 pKa = 4.01IFRR106 pKa = 11.84DD107 pKa = 3.73CRR109 pKa = 11.84LRR111 pKa = 11.84LALVLDD117 pKa = 4.71AACGCVDD124 pKa = 4.33CFRR127 pKa = 11.84LAKK130 pKa = 9.37VLKK133 pKa = 9.67VAPRR137 pKa = 11.84CLRR140 pKa = 11.84PPKK143 pKa = 10.56LMAHH147 pKa = 6.5EE148 pKa = 4.42RR149 pKa = 11.84QCAAASTLTYY159 pKa = 9.8FYY161 pKa = 10.66NSYY164 pKa = 9.43MVAGSDD170 pKa = 3.64VEE172 pKa = 4.3VAPEE176 pKa = 4.44HH177 pKa = 7.13IEE179 pKa = 3.98LLLIRR184 pKa = 11.84PDD186 pKa = 3.53QLNSYY191 pKa = 9.32PYY193 pKa = 10.38GWDD196 pKa = 3.35EE197 pKa = 4.04EE198 pKa = 4.9VAGLMSCLNLCWLYY212 pKa = 11.12TMMYY216 pKa = 8.73EE217 pKa = 3.84HH218 pKa = 7.57ATRR221 pKa = 11.84GMCTLTRR228 pKa = 11.84ALEE231 pKa = 4.07EE232 pKa = 4.22AYY234 pKa = 10.52HH235 pKa = 6.03RR236 pKa = 11.84VRR238 pKa = 11.84PGSKK242 pKa = 8.73ATLEE246 pKa = 4.07NMVQAVIEE254 pKa = 4.4GDD256 pKa = 3.49SRR258 pKa = 11.84ASRR261 pKa = 11.84SEE263 pKa = 3.75EE264 pKa = 3.97PRR266 pKa = 11.84PHH268 pKa = 7.22HH269 pKa = 5.55YY270 pKa = 9.61TLIKK274 pKa = 10.15PLVTMKK280 pKa = 10.62HH281 pKa = 3.82EE282 pKa = 4.51HH283 pKa = 6.13RR284 pKa = 11.84WSEE287 pKa = 4.12KK288 pKa = 9.53QDD290 pKa = 3.43SAVQTQVSCLYY301 pKa = 9.88EE302 pKa = 4.2LVKK305 pKa = 10.84GMYY308 pKa = 9.89GPYY311 pKa = 10.1ARR313 pKa = 11.84QVQYY317 pKa = 11.09GRR319 pKa = 11.84KK320 pKa = 6.81PTDD323 pKa = 3.28MVPPLLSSVLMGVHH337 pKa = 7.02PEE339 pKa = 3.93LLRR342 pKa = 11.84EE343 pKa = 4.18KK344 pKa = 10.42DD345 pKa = 3.61PPLCQPLQPPLGSSSATGDD364 pKa = 3.23KK365 pKa = 11.1AGDD368 pKa = 3.55VSAEE372 pKa = 3.9RR373 pKa = 11.84AAGRR377 pKa = 11.84ASEE380 pKa = 4.53AEE382 pKa = 4.01GDD384 pKa = 3.78RR385 pKa = 11.84QYY387 pKa = 8.06TTCGCLGACACGTLDD402 pKa = 3.46TTEE405 pKa = 4.28GEE407 pKa = 4.54IVYY410 pKa = 9.79HH411 pKa = 6.51AARR414 pKa = 11.84GTGPKK419 pKa = 9.48PFCLGTGSGQIPCRR433 pKa = 11.84GEE435 pKa = 3.94DD436 pKa = 3.29VRR438 pKa = 11.84DD439 pKa = 3.77ARR441 pKa = 11.84DD442 pKa = 3.53PTCQRR447 pKa = 11.84EE448 pKa = 4.56AIGEE452 pKa = 4.28VTEE455 pKa = 5.53GADD458 pKa = 3.97QVDD461 pKa = 3.75KK462 pKa = 8.99TTTQRR467 pKa = 11.84AASQFEE473 pKa = 4.27PTPEE477 pKa = 3.66DD478 pKa = 3.82VIRR481 pKa = 11.84LGARR485 pKa = 11.84RR486 pKa = 11.84KK487 pKa = 8.11TSGIGTAAGDD497 pKa = 3.68VLRR500 pKa = 11.84PDD502 pKa = 3.55IAQNQYY508 pKa = 10.69PLLALDD514 pKa = 4.25PRR516 pKa = 11.84DD517 pKa = 3.6LEE519 pKa = 4.3SRR521 pKa = 11.84RR522 pKa = 11.84VQHH525 pKa = 5.78VFSDD529 pKa = 3.72EE530 pKa = 4.16RR531 pKa = 11.84AGSPEE536 pKa = 3.89TGEE539 pKa = 4.28PDD541 pKa = 3.11QEE543 pKa = 3.93PRR545 pKa = 11.84KK546 pKa = 10.01RR547 pKa = 11.84KK548 pKa = 9.25PEE550 pKa = 4.2SGNGSPGASPFFKK563 pKa = 10.56SIWGGGDD570 pKa = 3.39YY571 pKa = 10.78VRR573 pKa = 11.84QSPPLGYY580 pKa = 10.82GPTKK584 pKa = 10.63DD585 pKa = 3.08RR586 pKa = 11.84AVGDD590 pKa = 3.97TYY592 pKa = 11.26WNYY595 pKa = 10.74TGTDD599 pKa = 3.72DD600 pKa = 4.19EE601 pKa = 4.76QEE603 pKa = 3.91RR604 pKa = 11.84CEE606 pKa = 4.05ADD608 pKa = 3.37GVGDD612 pKa = 3.52EE613 pKa = 6.04DD614 pKa = 4.73EE615 pKa = 5.97YY616 pKa = 11.5EE617 pKa = 4.64DD618 pKa = 5.86CDD620 pKa = 4.38EE621 pKa = 5.62LDD623 pKa = 4.61LRR625 pKa = 11.84DD626 pKa = 3.88TKK628 pKa = 10.87TGGCHH633 pKa = 5.3ATLGGDD639 pKa = 4.18GNQEE643 pKa = 3.92PPFLLLRR650 pKa = 11.84DD651 pKa = 4.56FEE653 pKa = 5.37ALLKK657 pKa = 9.79TGSSEE662 pKa = 3.82GLSPHH667 pKa = 6.04IQVSHH672 pKa = 5.23GTEE675 pKa = 3.97KK676 pKa = 10.09EE677 pKa = 4.17TLEE680 pKa = 5.59DD681 pKa = 3.88GGSCLKK687 pKa = 10.87VYY689 pKa = 10.71EE690 pKa = 5.01DD691 pKa = 4.25DD692 pKa = 4.48SKK694 pKa = 11.86GEE696 pKa = 4.1SGGNPRR702 pKa = 11.84DD703 pKa = 3.46VTGNVEE709 pKa = 4.27DD710 pKa = 5.35AVDD713 pKa = 5.0GYY715 pKa = 11.8NSDD718 pKa = 4.6DD719 pKa = 5.12LGISCCQDD727 pKa = 2.37GWSGDD732 pKa = 3.61DD733 pKa = 4.01GEE735 pKa = 5.58AVDD738 pKa = 5.59TEE740 pKa = 5.49DD741 pKa = 3.99EE742 pKa = 4.09DD743 pKa = 4.79TYY745 pKa = 11.51TNQEE749 pKa = 3.83GCGGGVEE756 pKa = 4.2EE757 pKa = 5.97GEE759 pKa = 3.96DD760 pKa = 3.25HH761 pKa = 7.78GIGEE765 pKa = 4.48EE766 pKa = 3.99EE767 pKa = 4.7DD768 pKa = 4.95GGDD771 pKa = 3.19EE772 pKa = 4.38CYY774 pKa = 10.51EE775 pKa = 4.23DD776 pKa = 4.66EE777 pKa = 4.8GAQSSYY783 pKa = 10.07YY784 pKa = 10.78DD785 pKa = 3.62RR786 pKa = 11.84VAGDD790 pKa = 3.4NADD793 pKa = 3.73EE794 pKa = 4.68LGEE797 pKa = 4.0EE798 pKa = 4.23LGEE801 pKa = 4.35DD802 pKa = 3.84DD803 pKa = 5.41EE804 pKa = 6.09LGEE807 pKa = 4.08EE808 pKa = 4.4LGDD811 pKa = 3.74EE812 pKa = 4.25EE813 pKa = 5.23SVEE816 pKa = 5.1DD817 pKa = 4.52EE818 pKa = 4.63DD819 pKa = 5.99SGDD822 pKa = 3.73GGDD825 pKa = 3.98GEE827 pKa = 4.98EE828 pKa = 4.02VCEE831 pKa = 4.98DD832 pKa = 4.06YY833 pKa = 10.9EE834 pKa = 4.52VCDD837 pKa = 3.78GDD839 pKa = 4.3EE840 pKa = 4.12EE841 pKa = 5.04CEE843 pKa = 4.12EE844 pKa = 4.52GNDD847 pKa = 3.57TGLEE851 pKa = 4.05TLEE854 pKa = 5.18DD855 pKa = 4.08SVDD858 pKa = 3.68EE859 pKa = 4.5EE860 pKa = 4.29NMGIYY865 pKa = 9.98FRR867 pKa = 11.84EE868 pKa = 4.59HH869 pKa = 5.35II870 pKa = 4.38
Molecular weight: 94.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D1A3K3|A0A2D1A3K3_9GAMA DNA polymerase processivity subunit OS=Eptesicus fuscus gammaherpesvirus OX=2035399 PE=4 SV=1
MM1 pKa = 7.11YY2 pKa = 9.06PRR4 pKa = 11.84RR5 pKa = 11.84LSRR8 pKa = 11.84QRR10 pKa = 11.84STLSVFRR17 pKa = 11.84FRR19 pKa = 11.84VTLIYY24 pKa = 10.51CDD26 pKa = 3.79VVDD29 pKa = 4.9PSSDD33 pKa = 2.97HH34 pKa = 6.93LRR36 pKa = 11.84FCISHH41 pKa = 6.95RR42 pKa = 11.84EE43 pKa = 3.92LSWRR47 pKa = 11.84SRR49 pKa = 11.84YY50 pKa = 8.98RR51 pKa = 11.84ACSDD55 pKa = 2.55SWKK58 pKa = 9.25TLTCSSGIRR67 pKa = 11.84CGCWPACLPPVANSVKK83 pKa = 9.95RR84 pKa = 11.84VSRR87 pKa = 11.84STTQHH92 pKa = 5.06ITTFTLDD99 pKa = 3.62KK100 pKa = 10.15GQHH103 pKa = 5.3SQLIPQLTPQARR115 pKa = 11.84CHH117 pKa = 6.38LVLITWGDD125 pKa = 3.57GGRR128 pKa = 11.84ATASSARR135 pKa = 11.84LPPTVIYY142 pKa = 8.66CQPFAKK148 pKa = 10.06RR149 pKa = 11.84AFRR152 pKa = 11.84HH153 pKa = 5.06LCRR156 pKa = 11.84MKK158 pKa = 10.88LIIFTHH164 pKa = 5.79MPFLIIILLFVLLQQ178 pKa = 3.59
MM1 pKa = 7.11YY2 pKa = 9.06PRR4 pKa = 11.84RR5 pKa = 11.84LSRR8 pKa = 11.84QRR10 pKa = 11.84STLSVFRR17 pKa = 11.84FRR19 pKa = 11.84VTLIYY24 pKa = 10.51CDD26 pKa = 3.79VVDD29 pKa = 4.9PSSDD33 pKa = 2.97HH34 pKa = 6.93LRR36 pKa = 11.84FCISHH41 pKa = 6.95RR42 pKa = 11.84EE43 pKa = 3.92LSWRR47 pKa = 11.84SRR49 pKa = 11.84YY50 pKa = 8.98RR51 pKa = 11.84ACSDD55 pKa = 2.55SWKK58 pKa = 9.25TLTCSSGIRR67 pKa = 11.84CGCWPACLPPVANSVKK83 pKa = 9.95RR84 pKa = 11.84VSRR87 pKa = 11.84STTQHH92 pKa = 5.06ITTFTLDD99 pKa = 3.62KK100 pKa = 10.15GQHH103 pKa = 5.3SQLIPQLTPQARR115 pKa = 11.84CHH117 pKa = 6.38LVLITWGDD125 pKa = 3.57GGRR128 pKa = 11.84ATASSARR135 pKa = 11.84LPPTVIYY142 pKa = 8.66CQPFAKK148 pKa = 10.06RR149 pKa = 11.84AFRR152 pKa = 11.84HH153 pKa = 5.06LCRR156 pKa = 11.84MKK158 pKa = 10.88LIIFTHH164 pKa = 5.79MPFLIIILLFVLLQQ178 pKa = 3.59
Molecular weight: 20.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
42028 |
89 |
3622 |
560.4 |
61.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.712 ± 0.269 | 2.184 ± 0.157 |
5.092 ± 0.186 | 5.703 ± 0.252 |
3.609 ± 0.188 | 7.321 ± 0.301 |
2.441 ± 0.093 | 3.086 ± 0.163 |
2.77 ± 0.139 | 9.648 ± 0.214 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.11 ± 0.102 | 2.936 ± 0.11 |
7.105 ± 0.294 | 3.89 ± 0.28 |
7.654 ± 0.242 | 7.371 ± 0.229 |
6.103 ± 0.307 | 6.489 ± 0.237 |
1.128 ± 0.076 | 2.646 ± 0.118 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
