
Synechococcus sp. MIT S9508
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; unclassified Synechococcus
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
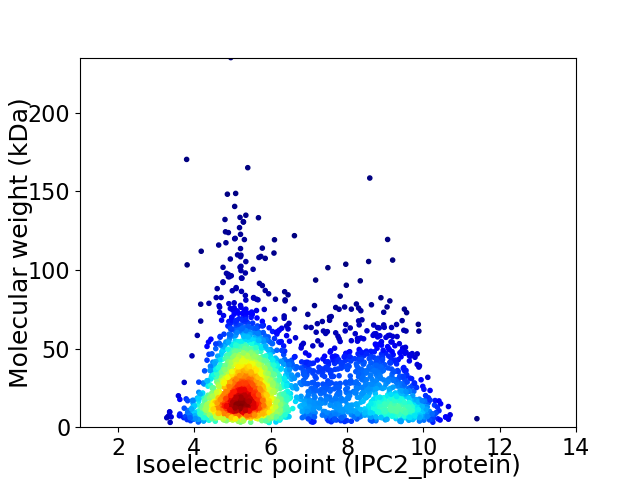
Virtual 2D-PAGE plot for 2817 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A164C9S5|A0A164C9S5_9SYNE Imidazole glycerol phosphate synthase subunit HisF OS=Synechococcus sp. MIT S9508 OX=1801629 GN=hisF PE=3 SV=1
MM1 pKa = 7.58TFTGQYY7 pKa = 11.09VNIQSHH13 pKa = 5.82SSGSQGSTTALRR25 pKa = 11.84NTSFIATSDD34 pKa = 3.07LGTRR38 pKa = 11.84IDD40 pKa = 4.41INASSNSNGGWWWNSGSSNTTGISNTSIEE69 pKa = 4.47TGSGNDD75 pKa = 3.71VININANANSNGGGWWWRR93 pKa = 11.84RR94 pKa = 11.84GGNADD99 pKa = 4.74SVGIDD104 pKa = 3.36NNSSINTGAGNDD116 pKa = 3.59SVYY119 pKa = 11.32VNAFASGAATNAWAIRR135 pKa = 11.84DD136 pKa = 3.65SSIDD140 pKa = 3.45VGSGDD145 pKa = 5.04DD146 pKa = 3.87VVSLNATTFQTQRR159 pKa = 11.84RR160 pKa = 11.84YY161 pKa = 10.67DD162 pKa = 3.83PAWGSEE168 pKa = 3.81NSSIDD173 pKa = 3.54LGSGNDD179 pKa = 3.2RR180 pKa = 11.84LFINANASGQGEE192 pKa = 4.08MRR194 pKa = 11.84AFGALDD200 pKa = 3.39STIDD204 pKa = 3.56AGSGDD209 pKa = 4.13DD210 pKa = 4.8AISINASANSNNGWWWWGGGNDD232 pKa = 3.67PATAKK237 pKa = 10.67GLANSDD243 pKa = 3.18VDD245 pKa = 4.47LGSGNDD251 pKa = 3.76FLSVSASTNGGISNATAVEE270 pKa = 4.29DD271 pKa = 3.77SSIDD275 pKa = 3.4AGSGNDD281 pKa = 3.59VININTNARR290 pKa = 11.84ANGGGWRR297 pKa = 11.84WWNRR301 pKa = 11.84GGNPATAKK309 pKa = 10.79GLDD312 pKa = 3.4NSEE315 pKa = 4.06ISTGDD320 pKa = 3.41GQDD323 pKa = 3.57SISINTTATSNGGGWWWGGGGNADD347 pKa = 4.71SVGIDD352 pKa = 3.36NNSSINTGADD362 pKa = 3.14NDD364 pKa = 4.29SVYY367 pKa = 11.37VNAFASGAATNAWAIRR383 pKa = 11.84DD384 pKa = 3.65SSIDD388 pKa = 3.45VGSGDD393 pKa = 5.04DD394 pKa = 3.87VVSLNATTFQTQRR407 pKa = 11.84RR408 pKa = 11.84YY409 pKa = 10.62DD410 pKa = 3.48PAYY413 pKa = 9.77GSEE416 pKa = 4.08NSSIDD421 pKa = 3.54LGSGNDD427 pKa = 3.2RR428 pKa = 11.84LFINANASGQGDD440 pKa = 3.1IRR442 pKa = 11.84AFGALDD448 pKa = 3.43TTIDD452 pKa = 3.75AGSGDD457 pKa = 4.13DD458 pKa = 4.8AISINASANNNNWGWGNAATATGIGSTEE486 pKa = 3.72ISTGSGDD493 pKa = 4.31DD494 pKa = 3.59SLSINATAHH503 pKa = 5.31GRR505 pKa = 11.84QNEE508 pKa = 4.12WSSQSSWGYY517 pKa = 10.38DD518 pKa = 3.32YY519 pKa = 10.99SYY521 pKa = 11.5SGATQWSSEE530 pKa = 4.03YY531 pKa = 10.71SHH533 pKa = 6.67SANLAFGGGWWGSSSLSYY551 pKa = 10.64GYY553 pKa = 9.57QSKK556 pKa = 10.9YY557 pKa = 11.05DD558 pKa = 3.67YY559 pKa = 10.71DD560 pKa = 3.76YY561 pKa = 11.0EE562 pKa = 4.31YY563 pKa = 11.4ANQGHH568 pKa = 6.26YY569 pKa = 10.04KK570 pKa = 10.15SSHH573 pKa = 4.92SSRR576 pKa = 11.84NVDD579 pKa = 2.72ISGGEE584 pKa = 3.92SYY586 pKa = 11.21GAINSSIDD594 pKa = 3.71LGSGDD599 pKa = 3.27NTATVNVLAGNQAVGFRR616 pKa = 11.84ATSLNSGNGDD626 pKa = 3.43DD627 pKa = 5.03SFSLSVNANGEE638 pKa = 3.84RR639 pKa = 11.84SYY641 pKa = 11.33SYY643 pKa = 10.86TSDD646 pKa = 3.5YY647 pKa = 11.17EE648 pKa = 4.43YY649 pKa = 11.23ASSGYY654 pKa = 10.55DD655 pKa = 2.66IGSNSGEE662 pKa = 4.02SSSWRR667 pKa = 11.84SYY669 pKa = 9.86QNNSRR674 pKa = 11.84AWWWGNSQSQSEE686 pKa = 4.72HH687 pKa = 5.67ASKK690 pKa = 10.34WDD692 pKa = 3.39NSWNHH697 pKa = 4.24SHH699 pKa = 6.94EE700 pKa = 4.25YY701 pKa = 9.64SNKK704 pKa = 9.64SASEE708 pKa = 3.84HH709 pKa = 7.13SNTQRR714 pKa = 11.84FGSSLGSEE722 pKa = 4.22NSTIDD727 pKa = 5.03LGNGDD732 pKa = 4.19NNAEE736 pKa = 3.92ISVKK740 pKa = 10.39GGEE743 pKa = 4.16RR744 pKa = 11.84ALGLISSDD752 pKa = 3.65LLTGSGSDD760 pKa = 3.76TISIDD765 pKa = 3.2TSAEE769 pKa = 3.8GLVEE773 pKa = 3.88YY774 pKa = 10.99SNTSKK779 pKa = 11.05GSNSNSYY786 pKa = 10.98SNIYY790 pKa = 10.16NYY792 pKa = 10.95SNSNSHH798 pKa = 6.44EE799 pKa = 3.93YY800 pKa = 10.39SYY802 pKa = 11.86SSARR806 pKa = 11.84WYY808 pKa = 10.63GHH810 pKa = 5.38NQSSSEE816 pKa = 3.75GNYY819 pKa = 10.04DD820 pKa = 3.5YY821 pKa = 11.27KK822 pKa = 11.38SQGEE826 pKa = 4.3YY827 pKa = 10.32TNQGQSNWDD836 pKa = 3.28NEE838 pKa = 3.99YY839 pKa = 10.76SRR841 pKa = 11.84TYY843 pKa = 10.73RR844 pKa = 11.84FGVSIGAEE852 pKa = 4.06DD853 pKa = 3.77SSIDD857 pKa = 4.17AGDD860 pKa = 3.84GNNSINLQSIGGEE873 pKa = 3.94LAIALAGSSITAGSGDD889 pKa = 4.31DD890 pKa = 3.41SLAIIGLAEE899 pKa = 4.75GEE901 pKa = 3.84DD902 pKa = 3.95SYY904 pKa = 12.06INRR907 pKa = 11.84KK908 pKa = 9.24KK909 pKa = 11.07SNSTWNHH916 pKa = 6.4DD917 pKa = 3.03SWNSNKK923 pKa = 9.72RR924 pKa = 11.84SEE926 pKa = 4.48EE927 pKa = 3.7YY928 pKa = 10.4SRR930 pKa = 11.84SYY932 pKa = 9.9TSAYY936 pKa = 8.42YY937 pKa = 8.88WYY939 pKa = 10.69NRR941 pKa = 11.84IEE943 pKa = 4.45EE944 pKa = 4.26YY945 pKa = 10.8SSNHH949 pKa = 5.32EE950 pKa = 4.23SEE952 pKa = 4.58SDD954 pKa = 3.5GFSISLSRR962 pKa = 11.84YY963 pKa = 6.15NQNYY967 pKa = 7.72EE968 pKa = 3.49YY969 pKa = 11.3SRR971 pKa = 11.84IKK973 pKa = 10.74RR974 pKa = 11.84FGTAYY979 pKa = 10.04GAQKK983 pKa = 10.87SSINLGNGDD992 pKa = 3.81NEE994 pKa = 4.29ASISANGGEE1003 pKa = 4.39SAEE1006 pKa = 4.18ALSDD1010 pKa = 3.73SSLEE1014 pKa = 4.09TGKK1017 pKa = 11.11GDD1019 pKa = 4.69DD1020 pKa = 4.52VISLSATALGEE1031 pKa = 3.8NSLINKK1037 pKa = 9.33NEE1039 pKa = 3.76YY1040 pKa = 10.33LSEE1043 pKa = 4.22HH1044 pKa = 6.4SSVYY1048 pKa = 8.35SHH1050 pKa = 6.95KK1051 pKa = 10.71SEE1053 pKa = 3.88YY1054 pKa = 10.32SYY1056 pKa = 11.04QGEE1059 pKa = 4.27GLNQSRR1065 pKa = 11.84YY1066 pKa = 8.86GWGWWNPTSEE1076 pKa = 4.76SSHH1079 pKa = 6.46SYY1081 pKa = 9.56NNKK1084 pKa = 9.61GNHH1087 pKa = 5.78NSQGNNQSLFIEE1099 pKa = 5.07DD1100 pKa = 4.35YY1101 pKa = 10.75EE1102 pKa = 4.32NSLIQKK1108 pKa = 9.51LGSAIGANRR1117 pKa = 11.84STIDD1121 pKa = 3.25TGSGDD1126 pKa = 4.33DD1127 pKa = 5.27LLTIDD1132 pKa = 3.81AKK1134 pKa = 11.21GYY1136 pKa = 8.57TEE1138 pKa = 4.92AVGIKK1143 pKa = 10.14NSSIDD1148 pKa = 3.58LGKK1151 pKa = 10.84GDD1153 pKa = 4.85DD1154 pKa = 3.73VFTISSSAFGFTSYY1168 pKa = 10.62LRR1170 pKa = 11.84QGLGSYY1176 pKa = 10.03DD1177 pKa = 3.38YY1178 pKa = 11.12SYY1180 pKa = 11.14DD1181 pKa = 3.8YY1182 pKa = 10.93QSSGSQSGEE1191 pKa = 3.49YY1192 pKa = 9.9SNSYY1196 pKa = 9.4RR1197 pKa = 11.84YY1198 pKa = 9.98SYY1200 pKa = 9.97PWWSYY1205 pKa = 8.11GRR1207 pKa = 11.84SYY1209 pKa = 10.79SGSYY1213 pKa = 9.54GSQWNYY1219 pKa = 9.9GYY1221 pKa = 11.03GYY1223 pKa = 10.12ASSGTWSYY1231 pKa = 12.11NEE1233 pKa = 4.57DD1234 pKa = 2.81SWLINTGIAKK1244 pKa = 10.15GAVNSTIDD1252 pKa = 3.35TGKK1255 pKa = 10.83GDD1257 pKa = 4.41DD1258 pKa = 4.79NISITARR1265 pKa = 11.84ASSKK1269 pKa = 9.6ATAIEE1274 pKa = 4.27DD1275 pKa = 4.17SEE1277 pKa = 4.72VLTGDD1282 pKa = 3.47GDD1284 pKa = 4.24DD1285 pKa = 3.86SVEE1288 pKa = 3.95VSAAAEE1294 pKa = 3.89EE1295 pKa = 4.32DD1296 pKa = 3.11ATAIEE1301 pKa = 4.6DD1302 pKa = 4.25SEE1304 pKa = 4.87VSTGDD1309 pKa = 3.15GDD1311 pKa = 4.8DD1312 pKa = 3.78SVEE1315 pKa = 3.95VSAAAEE1321 pKa = 3.89EE1322 pKa = 4.32DD1323 pKa = 3.11ATAIEE1328 pKa = 4.6DD1329 pKa = 4.25SEE1331 pKa = 4.86VSTGDD1336 pKa = 3.5GEE1338 pKa = 4.84DD1339 pKa = 3.85LIEE1342 pKa = 4.33VEE1344 pKa = 4.69STAEE1348 pKa = 4.05SSDD1351 pKa = 3.5EE1352 pKa = 4.11ASDD1355 pKa = 3.47EE1356 pKa = 4.33GVAKK1360 pKa = 10.7GLVNTTIDD1368 pKa = 4.04AGADD1372 pKa = 3.35DD1373 pKa = 5.45DD1374 pKa = 4.44ALLMEE1379 pKa = 4.74VTGEE1383 pKa = 3.92TLAIGMEE1390 pKa = 3.93NSTLNVGDD1398 pKa = 4.12SDD1400 pKa = 5.83DD1401 pKa = 4.6IIDD1404 pKa = 4.63VIVEE1408 pKa = 4.05ATNEE1412 pKa = 3.9EE1413 pKa = 4.23DD1414 pKa = 4.07GEE1416 pKa = 4.62AIGLSGSTISSGGGDD1431 pKa = 3.27DD1432 pKa = 4.13SIYY1435 pKa = 10.44VEE1437 pKa = 4.22VWGEE1441 pKa = 3.97SSAKK1445 pKa = 10.8AMVDD1449 pKa = 3.46SLIEE1453 pKa = 4.44AGDD1456 pKa = 3.73GDD1458 pKa = 5.27DD1459 pKa = 5.05YY1460 pKa = 11.3ITIDD1464 pKa = 4.22GEE1466 pKa = 4.55IEE1468 pKa = 4.08NSTIDD1473 pKa = 4.27AGDD1476 pKa = 3.76GDD1478 pKa = 5.1DD1479 pKa = 5.31VVDD1482 pKa = 5.71LYY1484 pKa = 11.16GTGDD1488 pKa = 3.27ATVQGGAGEE1497 pKa = 4.72DD1498 pKa = 3.82EE1499 pKa = 4.71LNGDD1503 pKa = 4.23EE1504 pKa = 6.03GNDD1507 pKa = 3.76TLLGGADD1514 pKa = 3.5NDD1516 pKa = 3.83ILNGYY1521 pKa = 8.79SGNDD1525 pKa = 3.38TLKK1528 pKa = 10.91GGEE1531 pKa = 4.43GDD1533 pKa = 5.07DD1534 pKa = 4.57ILDD1537 pKa = 4.3AGIGLDD1543 pKa = 3.81KK1544 pKa = 11.0LYY1546 pKa = 11.16GGDD1549 pKa = 4.31GEE1551 pKa = 5.03DD1552 pKa = 3.72TFILNLGEE1560 pKa = 4.67GYY1562 pKa = 8.87ATIYY1566 pKa = 10.91DD1567 pKa = 3.81FVIGEE1572 pKa = 4.93DD1573 pKa = 3.03ILEE1576 pKa = 4.32FSDD1579 pKa = 3.49MGDD1582 pKa = 2.39ITMEE1586 pKa = 4.12YY1587 pKa = 10.09KK1588 pKa = 10.69DD1589 pKa = 4.52KK1590 pKa = 11.61NSLFYY1595 pKa = 11.12SGTDD1599 pKa = 3.38YY1600 pKa = 11.42LGVANGVQLTRR1611 pKa = 11.84QEE1613 pKa = 4.69DD1614 pKa = 3.71EE1615 pKa = 4.32TFAA1618 pKa = 5.62
MM1 pKa = 7.58TFTGQYY7 pKa = 11.09VNIQSHH13 pKa = 5.82SSGSQGSTTALRR25 pKa = 11.84NTSFIATSDD34 pKa = 3.07LGTRR38 pKa = 11.84IDD40 pKa = 4.41INASSNSNGGWWWNSGSSNTTGISNTSIEE69 pKa = 4.47TGSGNDD75 pKa = 3.71VININANANSNGGGWWWRR93 pKa = 11.84RR94 pKa = 11.84GGNADD99 pKa = 4.74SVGIDD104 pKa = 3.36NNSSINTGAGNDD116 pKa = 3.59SVYY119 pKa = 11.32VNAFASGAATNAWAIRR135 pKa = 11.84DD136 pKa = 3.65SSIDD140 pKa = 3.45VGSGDD145 pKa = 5.04DD146 pKa = 3.87VVSLNATTFQTQRR159 pKa = 11.84RR160 pKa = 11.84YY161 pKa = 10.67DD162 pKa = 3.83PAWGSEE168 pKa = 3.81NSSIDD173 pKa = 3.54LGSGNDD179 pKa = 3.2RR180 pKa = 11.84LFINANASGQGEE192 pKa = 4.08MRR194 pKa = 11.84AFGALDD200 pKa = 3.39STIDD204 pKa = 3.56AGSGDD209 pKa = 4.13DD210 pKa = 4.8AISINASANSNNGWWWWGGGNDD232 pKa = 3.67PATAKK237 pKa = 10.67GLANSDD243 pKa = 3.18VDD245 pKa = 4.47LGSGNDD251 pKa = 3.76FLSVSASTNGGISNATAVEE270 pKa = 4.29DD271 pKa = 3.77SSIDD275 pKa = 3.4AGSGNDD281 pKa = 3.59VININTNARR290 pKa = 11.84ANGGGWRR297 pKa = 11.84WWNRR301 pKa = 11.84GGNPATAKK309 pKa = 10.79GLDD312 pKa = 3.4NSEE315 pKa = 4.06ISTGDD320 pKa = 3.41GQDD323 pKa = 3.57SISINTTATSNGGGWWWGGGGNADD347 pKa = 4.71SVGIDD352 pKa = 3.36NNSSINTGADD362 pKa = 3.14NDD364 pKa = 4.29SVYY367 pKa = 11.37VNAFASGAATNAWAIRR383 pKa = 11.84DD384 pKa = 3.65SSIDD388 pKa = 3.45VGSGDD393 pKa = 5.04DD394 pKa = 3.87VVSLNATTFQTQRR407 pKa = 11.84RR408 pKa = 11.84YY409 pKa = 10.62DD410 pKa = 3.48PAYY413 pKa = 9.77GSEE416 pKa = 4.08NSSIDD421 pKa = 3.54LGSGNDD427 pKa = 3.2RR428 pKa = 11.84LFINANASGQGDD440 pKa = 3.1IRR442 pKa = 11.84AFGALDD448 pKa = 3.43TTIDD452 pKa = 3.75AGSGDD457 pKa = 4.13DD458 pKa = 4.8AISINASANNNNWGWGNAATATGIGSTEE486 pKa = 3.72ISTGSGDD493 pKa = 4.31DD494 pKa = 3.59SLSINATAHH503 pKa = 5.31GRR505 pKa = 11.84QNEE508 pKa = 4.12WSSQSSWGYY517 pKa = 10.38DD518 pKa = 3.32YY519 pKa = 10.99SYY521 pKa = 11.5SGATQWSSEE530 pKa = 4.03YY531 pKa = 10.71SHH533 pKa = 6.67SANLAFGGGWWGSSSLSYY551 pKa = 10.64GYY553 pKa = 9.57QSKK556 pKa = 10.9YY557 pKa = 11.05DD558 pKa = 3.67YY559 pKa = 10.71DD560 pKa = 3.76YY561 pKa = 11.0EE562 pKa = 4.31YY563 pKa = 11.4ANQGHH568 pKa = 6.26YY569 pKa = 10.04KK570 pKa = 10.15SSHH573 pKa = 4.92SSRR576 pKa = 11.84NVDD579 pKa = 2.72ISGGEE584 pKa = 3.92SYY586 pKa = 11.21GAINSSIDD594 pKa = 3.71LGSGDD599 pKa = 3.27NTATVNVLAGNQAVGFRR616 pKa = 11.84ATSLNSGNGDD626 pKa = 3.43DD627 pKa = 5.03SFSLSVNANGEE638 pKa = 3.84RR639 pKa = 11.84SYY641 pKa = 11.33SYY643 pKa = 10.86TSDD646 pKa = 3.5YY647 pKa = 11.17EE648 pKa = 4.43YY649 pKa = 11.23ASSGYY654 pKa = 10.55DD655 pKa = 2.66IGSNSGEE662 pKa = 4.02SSSWRR667 pKa = 11.84SYY669 pKa = 9.86QNNSRR674 pKa = 11.84AWWWGNSQSQSEE686 pKa = 4.72HH687 pKa = 5.67ASKK690 pKa = 10.34WDD692 pKa = 3.39NSWNHH697 pKa = 4.24SHH699 pKa = 6.94EE700 pKa = 4.25YY701 pKa = 9.64SNKK704 pKa = 9.64SASEE708 pKa = 3.84HH709 pKa = 7.13SNTQRR714 pKa = 11.84FGSSLGSEE722 pKa = 4.22NSTIDD727 pKa = 5.03LGNGDD732 pKa = 4.19NNAEE736 pKa = 3.92ISVKK740 pKa = 10.39GGEE743 pKa = 4.16RR744 pKa = 11.84ALGLISSDD752 pKa = 3.65LLTGSGSDD760 pKa = 3.76TISIDD765 pKa = 3.2TSAEE769 pKa = 3.8GLVEE773 pKa = 3.88YY774 pKa = 10.99SNTSKK779 pKa = 11.05GSNSNSYY786 pKa = 10.98SNIYY790 pKa = 10.16NYY792 pKa = 10.95SNSNSHH798 pKa = 6.44EE799 pKa = 3.93YY800 pKa = 10.39SYY802 pKa = 11.86SSARR806 pKa = 11.84WYY808 pKa = 10.63GHH810 pKa = 5.38NQSSSEE816 pKa = 3.75GNYY819 pKa = 10.04DD820 pKa = 3.5YY821 pKa = 11.27KK822 pKa = 11.38SQGEE826 pKa = 4.3YY827 pKa = 10.32TNQGQSNWDD836 pKa = 3.28NEE838 pKa = 3.99YY839 pKa = 10.76SRR841 pKa = 11.84TYY843 pKa = 10.73RR844 pKa = 11.84FGVSIGAEE852 pKa = 4.06DD853 pKa = 3.77SSIDD857 pKa = 4.17AGDD860 pKa = 3.84GNNSINLQSIGGEE873 pKa = 3.94LAIALAGSSITAGSGDD889 pKa = 4.31DD890 pKa = 3.41SLAIIGLAEE899 pKa = 4.75GEE901 pKa = 3.84DD902 pKa = 3.95SYY904 pKa = 12.06INRR907 pKa = 11.84KK908 pKa = 9.24KK909 pKa = 11.07SNSTWNHH916 pKa = 6.4DD917 pKa = 3.03SWNSNKK923 pKa = 9.72RR924 pKa = 11.84SEE926 pKa = 4.48EE927 pKa = 3.7YY928 pKa = 10.4SRR930 pKa = 11.84SYY932 pKa = 9.9TSAYY936 pKa = 8.42YY937 pKa = 8.88WYY939 pKa = 10.69NRR941 pKa = 11.84IEE943 pKa = 4.45EE944 pKa = 4.26YY945 pKa = 10.8SSNHH949 pKa = 5.32EE950 pKa = 4.23SEE952 pKa = 4.58SDD954 pKa = 3.5GFSISLSRR962 pKa = 11.84YY963 pKa = 6.15NQNYY967 pKa = 7.72EE968 pKa = 3.49YY969 pKa = 11.3SRR971 pKa = 11.84IKK973 pKa = 10.74RR974 pKa = 11.84FGTAYY979 pKa = 10.04GAQKK983 pKa = 10.87SSINLGNGDD992 pKa = 3.81NEE994 pKa = 4.29ASISANGGEE1003 pKa = 4.39SAEE1006 pKa = 4.18ALSDD1010 pKa = 3.73SSLEE1014 pKa = 4.09TGKK1017 pKa = 11.11GDD1019 pKa = 4.69DD1020 pKa = 4.52VISLSATALGEE1031 pKa = 3.8NSLINKK1037 pKa = 9.33NEE1039 pKa = 3.76YY1040 pKa = 10.33LSEE1043 pKa = 4.22HH1044 pKa = 6.4SSVYY1048 pKa = 8.35SHH1050 pKa = 6.95KK1051 pKa = 10.71SEE1053 pKa = 3.88YY1054 pKa = 10.32SYY1056 pKa = 11.04QGEE1059 pKa = 4.27GLNQSRR1065 pKa = 11.84YY1066 pKa = 8.86GWGWWNPTSEE1076 pKa = 4.76SSHH1079 pKa = 6.46SYY1081 pKa = 9.56NNKK1084 pKa = 9.61GNHH1087 pKa = 5.78NSQGNNQSLFIEE1099 pKa = 5.07DD1100 pKa = 4.35YY1101 pKa = 10.75EE1102 pKa = 4.32NSLIQKK1108 pKa = 9.51LGSAIGANRR1117 pKa = 11.84STIDD1121 pKa = 3.25TGSGDD1126 pKa = 4.33DD1127 pKa = 5.27LLTIDD1132 pKa = 3.81AKK1134 pKa = 11.21GYY1136 pKa = 8.57TEE1138 pKa = 4.92AVGIKK1143 pKa = 10.14NSSIDD1148 pKa = 3.58LGKK1151 pKa = 10.84GDD1153 pKa = 4.85DD1154 pKa = 3.73VFTISSSAFGFTSYY1168 pKa = 10.62LRR1170 pKa = 11.84QGLGSYY1176 pKa = 10.03DD1177 pKa = 3.38YY1178 pKa = 11.12SYY1180 pKa = 11.14DD1181 pKa = 3.8YY1182 pKa = 10.93QSSGSQSGEE1191 pKa = 3.49YY1192 pKa = 9.9SNSYY1196 pKa = 9.4RR1197 pKa = 11.84YY1198 pKa = 9.98SYY1200 pKa = 9.97PWWSYY1205 pKa = 8.11GRR1207 pKa = 11.84SYY1209 pKa = 10.79SGSYY1213 pKa = 9.54GSQWNYY1219 pKa = 9.9GYY1221 pKa = 11.03GYY1223 pKa = 10.12ASSGTWSYY1231 pKa = 12.11NEE1233 pKa = 4.57DD1234 pKa = 2.81SWLINTGIAKK1244 pKa = 10.15GAVNSTIDD1252 pKa = 3.35TGKK1255 pKa = 10.83GDD1257 pKa = 4.41DD1258 pKa = 4.79NISITARR1265 pKa = 11.84ASSKK1269 pKa = 9.6ATAIEE1274 pKa = 4.27DD1275 pKa = 4.17SEE1277 pKa = 4.72VLTGDD1282 pKa = 3.47GDD1284 pKa = 4.24DD1285 pKa = 3.86SVEE1288 pKa = 3.95VSAAAEE1294 pKa = 3.89EE1295 pKa = 4.32DD1296 pKa = 3.11ATAIEE1301 pKa = 4.6DD1302 pKa = 4.25SEE1304 pKa = 4.87VSTGDD1309 pKa = 3.15GDD1311 pKa = 4.8DD1312 pKa = 3.78SVEE1315 pKa = 3.95VSAAAEE1321 pKa = 3.89EE1322 pKa = 4.32DD1323 pKa = 3.11ATAIEE1328 pKa = 4.6DD1329 pKa = 4.25SEE1331 pKa = 4.86VSTGDD1336 pKa = 3.5GEE1338 pKa = 4.84DD1339 pKa = 3.85LIEE1342 pKa = 4.33VEE1344 pKa = 4.69STAEE1348 pKa = 4.05SSDD1351 pKa = 3.5EE1352 pKa = 4.11ASDD1355 pKa = 3.47EE1356 pKa = 4.33GVAKK1360 pKa = 10.7GLVNTTIDD1368 pKa = 4.04AGADD1372 pKa = 3.35DD1373 pKa = 5.45DD1374 pKa = 4.44ALLMEE1379 pKa = 4.74VTGEE1383 pKa = 3.92TLAIGMEE1390 pKa = 3.93NSTLNVGDD1398 pKa = 4.12SDD1400 pKa = 5.83DD1401 pKa = 4.6IIDD1404 pKa = 4.63VIVEE1408 pKa = 4.05ATNEE1412 pKa = 3.9EE1413 pKa = 4.23DD1414 pKa = 4.07GEE1416 pKa = 4.62AIGLSGSTISSGGGDD1431 pKa = 3.27DD1432 pKa = 4.13SIYY1435 pKa = 10.44VEE1437 pKa = 4.22VWGEE1441 pKa = 3.97SSAKK1445 pKa = 10.8AMVDD1449 pKa = 3.46SLIEE1453 pKa = 4.44AGDD1456 pKa = 3.73GDD1458 pKa = 5.27DD1459 pKa = 5.05YY1460 pKa = 11.3ITIDD1464 pKa = 4.22GEE1466 pKa = 4.55IEE1468 pKa = 4.08NSTIDD1473 pKa = 4.27AGDD1476 pKa = 3.76GDD1478 pKa = 5.1DD1479 pKa = 5.31VVDD1482 pKa = 5.71LYY1484 pKa = 11.16GTGDD1488 pKa = 3.27ATVQGGAGEE1497 pKa = 4.72DD1498 pKa = 3.82EE1499 pKa = 4.71LNGDD1503 pKa = 4.23EE1504 pKa = 6.03GNDD1507 pKa = 3.76TLLGGADD1514 pKa = 3.5NDD1516 pKa = 3.83ILNGYY1521 pKa = 8.79SGNDD1525 pKa = 3.38TLKK1528 pKa = 10.91GGEE1531 pKa = 4.43GDD1533 pKa = 5.07DD1534 pKa = 4.57ILDD1537 pKa = 4.3AGIGLDD1543 pKa = 3.81KK1544 pKa = 11.0LYY1546 pKa = 11.16GGDD1549 pKa = 4.31GEE1551 pKa = 5.03DD1552 pKa = 3.72TFILNLGEE1560 pKa = 4.67GYY1562 pKa = 8.87ATIYY1566 pKa = 10.91DD1567 pKa = 3.81FVIGEE1572 pKa = 4.93DD1573 pKa = 3.03ILEE1576 pKa = 4.32FSDD1579 pKa = 3.49MGDD1582 pKa = 2.39ITMEE1586 pKa = 4.12YY1587 pKa = 10.09KK1588 pKa = 10.69DD1589 pKa = 4.52KK1590 pKa = 11.61NSLFYY1595 pKa = 11.12SGTDD1599 pKa = 3.38YY1600 pKa = 11.42LGVANGVQLTRR1611 pKa = 11.84QEE1613 pKa = 4.69DD1614 pKa = 3.71EE1615 pKa = 4.32TFAA1618 pKa = 5.62
Molecular weight: 170.38 kDa
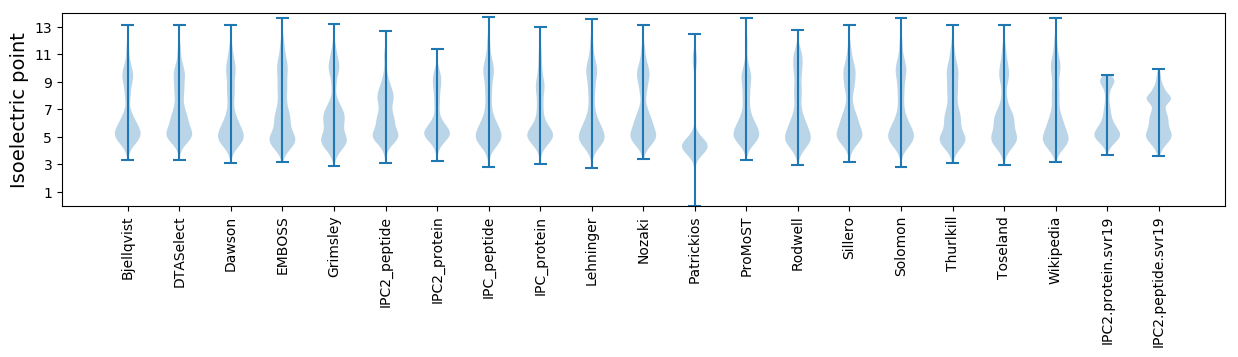
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A164CU86|A0A164CU86_9SYNE Uncharacterized protein OS=Synechococcus sp. MIT S9508 OX=1801629 GN=MITS9508_00087 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 8.87RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84THH26 pKa = 5.95TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.64RR38 pKa = 11.84GRR40 pKa = 11.84SRR42 pKa = 11.84LSVV45 pKa = 3.08
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 8.87RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84THH26 pKa = 5.95TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.64RR38 pKa = 11.84GRR40 pKa = 11.84SRR42 pKa = 11.84LSVV45 pKa = 3.08
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
737073 |
29 |
2175 |
261.7 |
28.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.825 ± 0.062 | 1.293 ± 0.017 |
5.447 ± 0.041 | 5.857 ± 0.048 |
3.183 ± 0.039 | 8.002 ± 0.046 |
2.077 ± 0.025 | 4.577 ± 0.039 |
3.015 ± 0.042 | 12.312 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.15 ± 0.021 | 2.859 ± 0.036 |
5.228 ± 0.036 | 4.786 ± 0.036 |
7.062 ± 0.051 | 6.904 ± 0.046 |
4.762 ± 0.034 | 6.99 ± 0.046 |
1.752 ± 0.033 | 1.918 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
