
Amycolatopsis marina
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Amycolatopsis
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
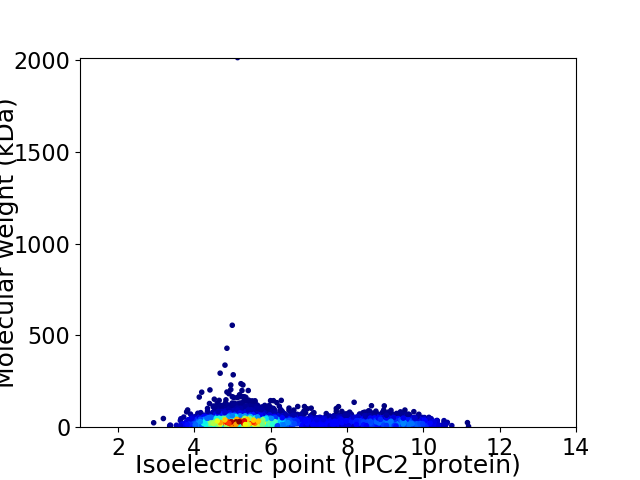
Virtual 2D-PAGE plot for 6643 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
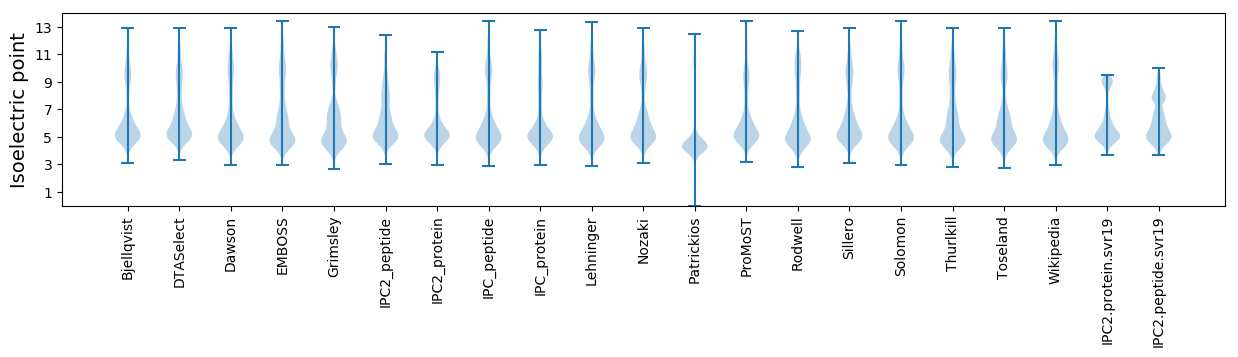
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1CQQ6|A0A1I1CQQ6_9PSEU DDE superfamily endonuclease (Fragment) OS=Amycolatopsis marina OX=490629 GN=SAMN05216266_1341 PE=4 SV=1
GG1 pKa = 7.29DD2 pKa = 3.42QDD4 pKa = 5.33AEE6 pKa = 4.26GNYY9 pKa = 9.57IGSATVTVTATDD21 pKa = 3.56ADD23 pKa = 4.13SGVDD27 pKa = 3.37TVEE30 pKa = 3.91YY31 pKa = 10.55QLDD34 pKa = 3.59GGAYY38 pKa = 8.5QAYY41 pKa = 5.56TTPITVDD48 pKa = 2.93TAGEE52 pKa = 3.91HH53 pKa = 5.08TLQYY57 pKa = 10.77RR58 pKa = 11.84ATDD61 pKa = 3.4NSGNTSEE68 pKa = 4.33AGSVIFTVTEE78 pKa = 3.94PAPDD82 pKa = 4.55DD83 pKa = 3.59STAPEE88 pKa = 3.93VSGEE92 pKa = 4.04VSGDD96 pKa = 2.97QDD98 pKa = 4.45AEE100 pKa = 4.21GNYY103 pKa = 9.94VGSATVTVTATDD115 pKa = 3.69ADD117 pKa = 4.02SGLDD121 pKa = 3.5TVHH124 pKa = 6.76FAIDD128 pKa = 3.47GGSYY132 pKa = 10.71NPYY135 pKa = 8.88TEE137 pKa = 5.31PIVVSEE143 pKa = 4.3PGEE146 pKa = 4.08HH147 pKa = 4.89TVSFRR152 pKa = 11.84ATDD155 pKa = 3.49NAGNTSEE162 pKa = 4.09IASVSFTVVAEE173 pKa = 4.83DD174 pKa = 4.78PDD176 pKa = 4.12DD177 pKa = 3.9TAPPQVNAEE186 pKa = 4.11VTGDD190 pKa = 3.25QDD192 pKa = 4.86AEE194 pKa = 4.26GNYY197 pKa = 9.94VGSATVTLSASDD209 pKa = 3.35TGSGVFALRR218 pKa = 11.84YY219 pKa = 9.72SLDD222 pKa = 3.7GGSFTPYY229 pKa = 10.51DD230 pKa = 4.07DD231 pKa = 4.92PLVLTAPGEE240 pKa = 4.22HH241 pKa = 5.31TVLYY245 pKa = 10.25RR246 pKa = 11.84ATDD249 pKa = 3.31NAGNVSEE256 pKa = 4.46TGSLTFTVVASDD268 pKa = 3.95SDD270 pKa = 3.95ACPGSDD276 pKa = 3.3VRR278 pKa = 11.84EE279 pKa = 3.95TVIIGNNDD287 pKa = 3.11STVANVDD294 pKa = 3.36TGDD297 pKa = 3.45GCTINDD303 pKa = 4.85LIDD306 pKa = 3.86EE307 pKa = 4.48NGEE310 pKa = 4.07YY311 pKa = 10.68ANHH314 pKa = 6.02GKK316 pKa = 9.4FVKK319 pKa = 10.01HH320 pKa = 5.22VRR322 pKa = 11.84QVTDD326 pKa = 3.49ALVADD331 pKa = 5.79GIISDD336 pKa = 3.94QEE338 pKa = 3.79KK339 pKa = 10.33GRR341 pKa = 11.84IMNAAARR348 pKa = 11.84SDD350 pKa = 3.5VGKK353 pKa = 10.72
GG1 pKa = 7.29DD2 pKa = 3.42QDD4 pKa = 5.33AEE6 pKa = 4.26GNYY9 pKa = 9.57IGSATVTVTATDD21 pKa = 3.56ADD23 pKa = 4.13SGVDD27 pKa = 3.37TVEE30 pKa = 3.91YY31 pKa = 10.55QLDD34 pKa = 3.59GGAYY38 pKa = 8.5QAYY41 pKa = 5.56TTPITVDD48 pKa = 2.93TAGEE52 pKa = 3.91HH53 pKa = 5.08TLQYY57 pKa = 10.77RR58 pKa = 11.84ATDD61 pKa = 3.4NSGNTSEE68 pKa = 4.33AGSVIFTVTEE78 pKa = 3.94PAPDD82 pKa = 4.55DD83 pKa = 3.59STAPEE88 pKa = 3.93VSGEE92 pKa = 4.04VSGDD96 pKa = 2.97QDD98 pKa = 4.45AEE100 pKa = 4.21GNYY103 pKa = 9.94VGSATVTVTATDD115 pKa = 3.69ADD117 pKa = 4.02SGLDD121 pKa = 3.5TVHH124 pKa = 6.76FAIDD128 pKa = 3.47GGSYY132 pKa = 10.71NPYY135 pKa = 8.88TEE137 pKa = 5.31PIVVSEE143 pKa = 4.3PGEE146 pKa = 4.08HH147 pKa = 4.89TVSFRR152 pKa = 11.84ATDD155 pKa = 3.49NAGNTSEE162 pKa = 4.09IASVSFTVVAEE173 pKa = 4.83DD174 pKa = 4.78PDD176 pKa = 4.12DD177 pKa = 3.9TAPPQVNAEE186 pKa = 4.11VTGDD190 pKa = 3.25QDD192 pKa = 4.86AEE194 pKa = 4.26GNYY197 pKa = 9.94VGSATVTLSASDD209 pKa = 3.35TGSGVFALRR218 pKa = 11.84YY219 pKa = 9.72SLDD222 pKa = 3.7GGSFTPYY229 pKa = 10.51DD230 pKa = 4.07DD231 pKa = 4.92PLVLTAPGEE240 pKa = 4.22HH241 pKa = 5.31TVLYY245 pKa = 10.25RR246 pKa = 11.84ATDD249 pKa = 3.31NAGNVSEE256 pKa = 4.46TGSLTFTVVASDD268 pKa = 3.95SDD270 pKa = 3.95ACPGSDD276 pKa = 3.3VRR278 pKa = 11.84EE279 pKa = 3.95TVIIGNNDD287 pKa = 3.11STVANVDD294 pKa = 3.36TGDD297 pKa = 3.45GCTINDD303 pKa = 4.85LIDD306 pKa = 3.86EE307 pKa = 4.48NGEE310 pKa = 4.07YY311 pKa = 10.68ANHH314 pKa = 6.02GKK316 pKa = 9.4FVKK319 pKa = 10.01HH320 pKa = 5.22VRR322 pKa = 11.84QVTDD326 pKa = 3.49ALVADD331 pKa = 5.79GIISDD336 pKa = 3.94QEE338 pKa = 3.79KK339 pKa = 10.33GRR341 pKa = 11.84IMNAAARR348 pKa = 11.84SDD350 pKa = 3.5VGKK353 pKa = 10.72
Molecular weight: 36.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0XF82|A0A1I0XF82_9PSEU Two-component system OmpR family sensor histidine kinase SenX3 OS=Amycolatopsis marina OX=490629 GN=SAMN05216266_103167 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.71QGKK33 pKa = 8.65
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.71QGKK33 pKa = 8.65
Molecular weight: 4.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2101265 |
27 |
18730 |
316.3 |
33.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.959 ± 0.039 | 0.782 ± 0.011 |
5.982 ± 0.027 | 5.95 ± 0.032 |
2.831 ± 0.019 | 9.221 ± 0.028 |
2.229 ± 0.014 | 3.566 ± 0.019 |
1.942 ± 0.018 | 10.485 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.773 ± 0.013 | 1.981 ± 0.016 |
5.73 ± 0.031 | 2.925 ± 0.019 |
7.944 ± 0.033 | 5.385 ± 0.02 |
6.006 ± 0.021 | 8.817 ± 0.029 |
1.489 ± 0.013 | 1.999 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
