
Bradymonas sediminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Bradymonadales; Bradymonadaceae; Bradymonas
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
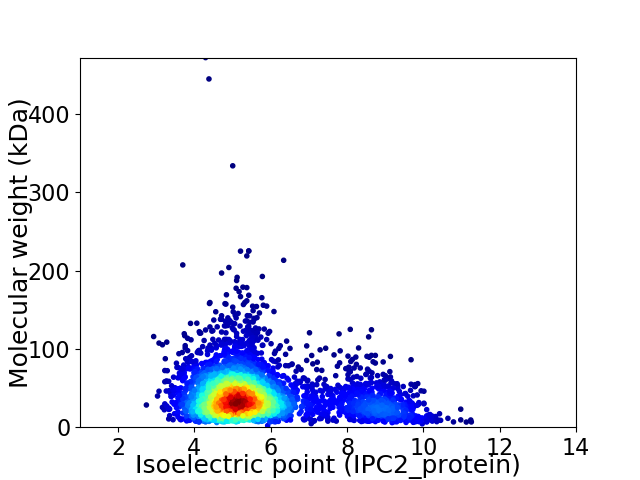
Virtual 2D-PAGE plot for 3697 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4FJB3|A0A2Z4FJB3_9DELT Uncharacterized protein OS=Bradymonas sediminis OX=1548548 GN=DN745_06080 PE=4 SV=1
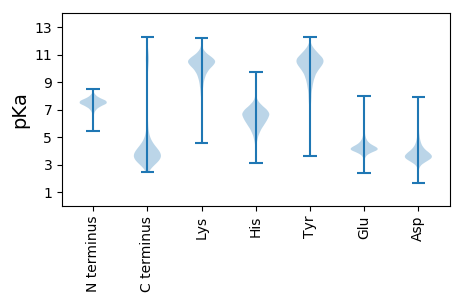
MM1 pKa = 7.79SDD3 pKa = 3.83SEE5 pKa = 4.71CPTDD9 pKa = 4.38RR10 pKa = 11.84VCGDD14 pKa = 3.57EE15 pKa = 4.61GVCVAPDD22 pKa = 4.05DD23 pKa = 4.57EE24 pKa = 6.03PDD26 pKa = 4.7AGDD29 pKa = 4.13VDD31 pKa = 5.47NNDD34 pKa = 3.73PDD36 pKa = 5.78AGDD39 pKa = 3.81VDD41 pKa = 5.26NNDD44 pKa = 3.89PDD46 pKa = 4.31GGSDD50 pKa = 3.75TLCPAPTITPAGAQNDD66 pKa = 4.09SSSASALPGSTIRR79 pKa = 11.84LDD81 pKa = 3.46GRR83 pKa = 11.84RR84 pKa = 11.84DD85 pKa = 3.32DD86 pKa = 4.71DD87 pKa = 5.22AVVTYY92 pKa = 7.45QWRR95 pKa = 11.84FVDD98 pKa = 5.2FPYY101 pKa = 10.75SEE103 pKa = 4.83TIAEE107 pKa = 4.46LEE109 pKa = 4.32PKK111 pKa = 10.31LVAHH115 pKa = 7.17ANGTADD121 pKa = 4.96FKK123 pKa = 11.78AEE125 pKa = 3.86ALGLYY130 pKa = 8.33TVEE133 pKa = 5.92LEE135 pKa = 4.41VKK137 pKa = 9.49DD138 pKa = 4.23ANGASVCPPTTAQVLVEE155 pKa = 4.57TDD157 pKa = 3.22DD158 pKa = 3.64QIYY161 pKa = 10.28VEE163 pKa = 5.15LVWEE167 pKa = 4.7TPGQTDD173 pKa = 3.67KK174 pKa = 11.41DD175 pKa = 3.64ASGPDD180 pKa = 3.37LEE182 pKa = 5.33LHH184 pKa = 5.95YY185 pKa = 10.47KK186 pKa = 9.5HH187 pKa = 7.14PNGTWAQWDD196 pKa = 3.93PPLDD200 pKa = 3.59IFWYY204 pKa = 10.59VPTADD209 pKa = 3.33WGIPGDD215 pKa = 4.09SSDD218 pKa = 4.15DD219 pKa = 3.39PVMRR223 pKa = 11.84RR224 pKa = 11.84FIDD227 pKa = 3.52HH228 pKa = 6.83GPGPEE233 pKa = 4.38AIAHH237 pKa = 6.16KK238 pKa = 10.53KK239 pKa = 10.11ADD241 pKa = 3.85PLVYY245 pKa = 10.34SVGVYY250 pKa = 10.33SYY252 pKa = 11.11DD253 pKa = 3.88DD254 pKa = 3.72KK255 pKa = 11.95DD256 pKa = 4.01FGPSFATVRR265 pKa = 11.84VYY267 pKa = 11.34LDD269 pKa = 3.69GEE271 pKa = 4.39LADD274 pKa = 3.9EE275 pKa = 4.55VPNIEE280 pKa = 4.24MPEE283 pKa = 4.08TNHH286 pKa = 6.09FYY288 pKa = 10.91EE289 pKa = 4.77VLTIDD294 pKa = 4.87AGQQTTEE301 pKa = 4.02YY302 pKa = 10.4LDD304 pKa = 3.27NHH306 pKa = 6.36YY307 pKa = 11.24EE308 pKa = 4.44DD309 pKa = 4.45FLEE312 pKa = 4.21MM313 pKa = 5.02
MM1 pKa = 7.79SDD3 pKa = 3.83SEE5 pKa = 4.71CPTDD9 pKa = 4.38RR10 pKa = 11.84VCGDD14 pKa = 3.57EE15 pKa = 4.61GVCVAPDD22 pKa = 4.05DD23 pKa = 4.57EE24 pKa = 6.03PDD26 pKa = 4.7AGDD29 pKa = 4.13VDD31 pKa = 5.47NNDD34 pKa = 3.73PDD36 pKa = 5.78AGDD39 pKa = 3.81VDD41 pKa = 5.26NNDD44 pKa = 3.89PDD46 pKa = 4.31GGSDD50 pKa = 3.75TLCPAPTITPAGAQNDD66 pKa = 4.09SSSASALPGSTIRR79 pKa = 11.84LDD81 pKa = 3.46GRR83 pKa = 11.84RR84 pKa = 11.84DD85 pKa = 3.32DD86 pKa = 4.71DD87 pKa = 5.22AVVTYY92 pKa = 7.45QWRR95 pKa = 11.84FVDD98 pKa = 5.2FPYY101 pKa = 10.75SEE103 pKa = 4.83TIAEE107 pKa = 4.46LEE109 pKa = 4.32PKK111 pKa = 10.31LVAHH115 pKa = 7.17ANGTADD121 pKa = 4.96FKK123 pKa = 11.78AEE125 pKa = 3.86ALGLYY130 pKa = 8.33TVEE133 pKa = 5.92LEE135 pKa = 4.41VKK137 pKa = 9.49DD138 pKa = 4.23ANGASVCPPTTAQVLVEE155 pKa = 4.57TDD157 pKa = 3.22DD158 pKa = 3.64QIYY161 pKa = 10.28VEE163 pKa = 5.15LVWEE167 pKa = 4.7TPGQTDD173 pKa = 3.67KK174 pKa = 11.41DD175 pKa = 3.64ASGPDD180 pKa = 3.37LEE182 pKa = 5.33LHH184 pKa = 5.95YY185 pKa = 10.47KK186 pKa = 9.5HH187 pKa = 7.14PNGTWAQWDD196 pKa = 3.93PPLDD200 pKa = 3.59IFWYY204 pKa = 10.59VPTADD209 pKa = 3.33WGIPGDD215 pKa = 4.09SSDD218 pKa = 4.15DD219 pKa = 3.39PVMRR223 pKa = 11.84RR224 pKa = 11.84FIDD227 pKa = 3.52HH228 pKa = 6.83GPGPEE233 pKa = 4.38AIAHH237 pKa = 6.16KK238 pKa = 10.53KK239 pKa = 10.11ADD241 pKa = 3.85PLVYY245 pKa = 10.34SVGVYY250 pKa = 10.33SYY252 pKa = 11.11DD253 pKa = 3.88DD254 pKa = 3.72KK255 pKa = 11.95DD256 pKa = 4.01FGPSFATVRR265 pKa = 11.84VYY267 pKa = 11.34LDD269 pKa = 3.69GEE271 pKa = 4.39LADD274 pKa = 3.9EE275 pKa = 4.55VPNIEE280 pKa = 4.24MPEE283 pKa = 4.08TNHH286 pKa = 6.09FYY288 pKa = 10.91EE289 pKa = 4.77VLTIDD294 pKa = 4.87AGQQTTEE301 pKa = 4.02YY302 pKa = 10.4LDD304 pKa = 3.27NHH306 pKa = 6.36YY307 pKa = 11.24EE308 pKa = 4.44DD309 pKa = 4.45FLEE312 pKa = 4.21MM313 pKa = 5.02
Molecular weight: 34.17 kDa
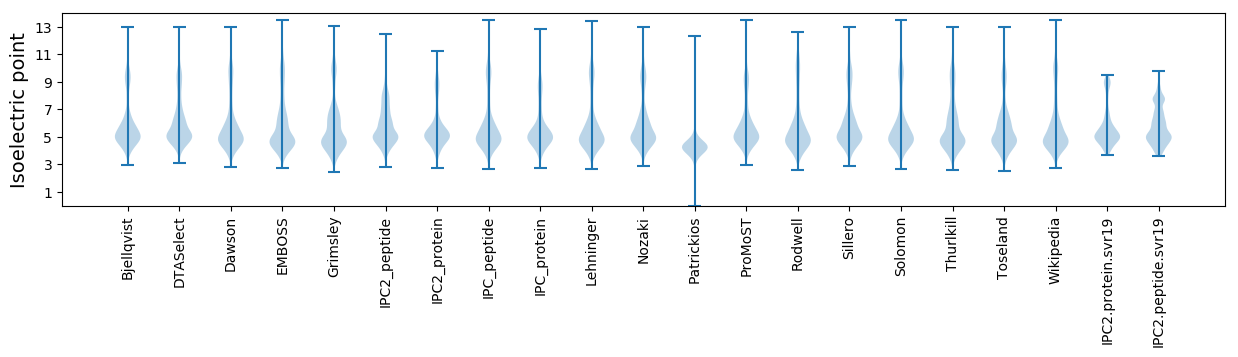
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4FLK3|A0A2Z4FLK3_9DELT Uncharacterized protein OS=Bradymonas sediminis OX=1548548 GN=DN745_10170 PE=4 SV=1
MM1 pKa = 7.08TRR3 pKa = 11.84TRR5 pKa = 11.84ALPPTPTTPPTPTPATRR22 pKa = 11.84PTPATRR28 pKa = 11.84PTPTTPPTPTTPPTPTTPRR47 pKa = 11.84AARR50 pKa = 11.84RR51 pKa = 11.84TVLTPSPWKK60 pKa = 8.95TPPSGPPTRR69 pKa = 11.84LRR71 pKa = 11.84RR72 pKa = 11.84PFWAKK77 pKa = 8.87LTARR81 pKa = 11.84AKK83 pKa = 10.08PAAA86 pKa = 4.19
MM1 pKa = 7.08TRR3 pKa = 11.84TRR5 pKa = 11.84ALPPTPTTPPTPTPATRR22 pKa = 11.84PTPATRR28 pKa = 11.84PTPTTPPTPTTPPTPTTPRR47 pKa = 11.84AARR50 pKa = 11.84RR51 pKa = 11.84TVLTPSPWKK60 pKa = 8.95TPPSGPPTRR69 pKa = 11.84LRR71 pKa = 11.84RR72 pKa = 11.84PFWAKK77 pKa = 8.87LTARR81 pKa = 11.84AKK83 pKa = 10.08PAAA86 pKa = 4.19
Molecular weight: 9.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1449336 |
18 |
4148 |
392.0 |
42.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.932 ± 0.054 | 1.185 ± 0.029 |
6.555 ± 0.036 | 6.998 ± 0.048 |
3.786 ± 0.023 | 7.922 ± 0.043 |
2.003 ± 0.018 | 4.961 ± 0.028 |
3.661 ± 0.032 | 9.706 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.239 ± 0.018 | 3.056 ± 0.024 |
4.984 ± 0.028 | 3.608 ± 0.022 |
6.467 ± 0.039 | 6.1 ± 0.027 |
5.107 ± 0.028 | 6.927 ± 0.031 |
1.241 ± 0.015 | 2.56 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
