
Changjiang polero-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
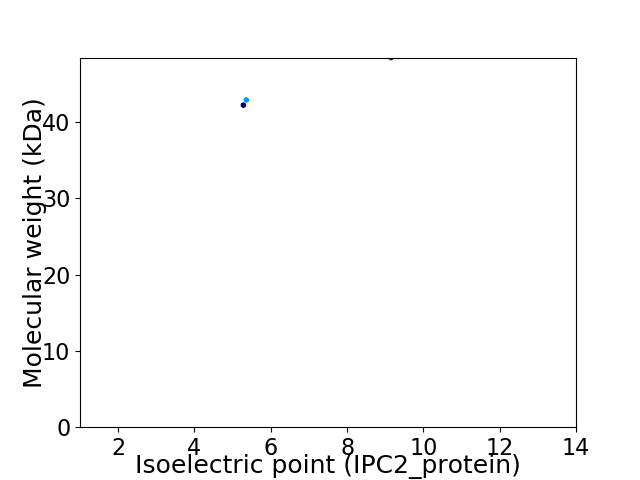
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KED2|A0A1L3KED2_9VIRU RNA-directed RNA polymerase OS=Changjiang polero-like virus 1 OX=1922799 PE=4 SV=1
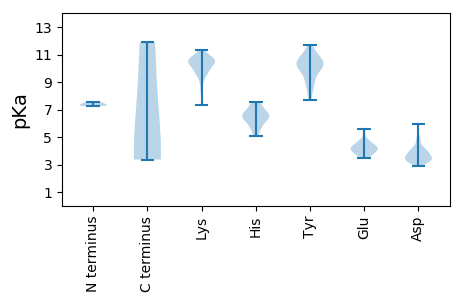
MM1 pKa = 7.26NSSYY5 pKa = 10.62IDD7 pKa = 3.92SRR9 pKa = 11.84PLSYY13 pKa = 11.64VNMYY17 pKa = 9.74KK18 pKa = 9.63WEE20 pKa = 4.19DD21 pKa = 3.86EE22 pKa = 4.17KK23 pKa = 11.08WSTVHH28 pKa = 6.24LTAGYY33 pKa = 9.78SRR35 pKa = 11.84NDD37 pKa = 3.51GEE39 pKa = 4.8EE40 pKa = 3.67IQPYY44 pKa = 8.87MVIPASKK51 pKa = 10.44GKK53 pKa = 9.76FRR55 pKa = 11.84VYY57 pKa = 10.42LEE59 pKa = 4.72CEE61 pKa = 3.53GFMAVKK67 pKa = 10.45SIGGKK72 pKa = 10.0ADD74 pKa = 3.55GCWAGLMAYY83 pKa = 10.17DD84 pKa = 4.05STKK87 pKa = 10.77SGWSVSEE94 pKa = 3.82YY95 pKa = 10.91VGAKK99 pKa = 7.34ITNIQANTNFVCGHH113 pKa = 7.31PDD115 pKa = 3.19VKK117 pKa = 11.3LNDD120 pKa = 3.5CTFKK124 pKa = 10.8KK125 pKa = 10.53NRR127 pKa = 11.84GIEE130 pKa = 4.03CDD132 pKa = 4.16FVCSFHH138 pKa = 7.53LDD140 pKa = 3.49CDD142 pKa = 4.23DD143 pKa = 5.97EE144 pKa = 5.57DD145 pKa = 4.79GKK147 pKa = 10.28WLLYY151 pKa = 9.2PPPVQKK157 pKa = 10.79SDD159 pKa = 3.39DD160 pKa = 3.54FNYY163 pKa = 9.85IVSYY167 pKa = 11.69SNFTDD172 pKa = 3.94KK173 pKa = 10.45WLEE176 pKa = 3.55WGMISISIDD185 pKa = 3.64EE186 pKa = 4.49INSPSSLQGLPRR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84LTQQLTTPEE209 pKa = 4.51GQPDD213 pKa = 3.92TNPAIGEE220 pKa = 4.27QATGPAPILSPNPALGEE237 pKa = 4.09EE238 pKa = 4.29KK239 pKa = 10.76LEE241 pKa = 4.73SIQRR245 pKa = 11.84WLSFSPSDD253 pKa = 3.93SEE255 pKa = 4.3QDD257 pKa = 2.97IPAPPDD263 pKa = 3.4PEE265 pKa = 5.37RR266 pKa = 11.84PTLPQEE272 pKa = 4.45VFRR275 pKa = 11.84VPPSNSIPSVIAPSLAGGSLRR296 pKa = 11.84GSLFGGSLKK305 pKa = 10.3PRR307 pKa = 11.84SLDD310 pKa = 3.39SGKK313 pKa = 10.58LPEE316 pKa = 4.21TTPRR320 pKa = 11.84DD321 pKa = 3.82NVSVAGSLTGSLVGGSLRR339 pKa = 11.84GGALKK344 pKa = 10.54SPRR347 pKa = 11.84LDD349 pKa = 2.97TAQAKK354 pKa = 8.46EE355 pKa = 3.75HH356 pKa = 6.35ARR358 pKa = 11.84ILKK361 pKa = 9.93QLGKK365 pKa = 8.1TAAAEE370 pKa = 3.8YY371 pKa = 10.15LEE373 pKa = 4.43RR374 pKa = 11.84VRR376 pKa = 11.84ASQSSGQDD384 pKa = 3.09FKK386 pKa = 11.88
MM1 pKa = 7.26NSSYY5 pKa = 10.62IDD7 pKa = 3.92SRR9 pKa = 11.84PLSYY13 pKa = 11.64VNMYY17 pKa = 9.74KK18 pKa = 9.63WEE20 pKa = 4.19DD21 pKa = 3.86EE22 pKa = 4.17KK23 pKa = 11.08WSTVHH28 pKa = 6.24LTAGYY33 pKa = 9.78SRR35 pKa = 11.84NDD37 pKa = 3.51GEE39 pKa = 4.8EE40 pKa = 3.67IQPYY44 pKa = 8.87MVIPASKK51 pKa = 10.44GKK53 pKa = 9.76FRR55 pKa = 11.84VYY57 pKa = 10.42LEE59 pKa = 4.72CEE61 pKa = 3.53GFMAVKK67 pKa = 10.45SIGGKK72 pKa = 10.0ADD74 pKa = 3.55GCWAGLMAYY83 pKa = 10.17DD84 pKa = 4.05STKK87 pKa = 10.77SGWSVSEE94 pKa = 3.82YY95 pKa = 10.91VGAKK99 pKa = 7.34ITNIQANTNFVCGHH113 pKa = 7.31PDD115 pKa = 3.19VKK117 pKa = 11.3LNDD120 pKa = 3.5CTFKK124 pKa = 10.8KK125 pKa = 10.53NRR127 pKa = 11.84GIEE130 pKa = 4.03CDD132 pKa = 4.16FVCSFHH138 pKa = 7.53LDD140 pKa = 3.49CDD142 pKa = 4.23DD143 pKa = 5.97EE144 pKa = 5.57DD145 pKa = 4.79GKK147 pKa = 10.28WLLYY151 pKa = 9.2PPPVQKK157 pKa = 10.79SDD159 pKa = 3.39DD160 pKa = 3.54FNYY163 pKa = 9.85IVSYY167 pKa = 11.69SNFTDD172 pKa = 3.94KK173 pKa = 10.45WLEE176 pKa = 3.55WGMISISIDD185 pKa = 3.64EE186 pKa = 4.49INSPSSLQGLPRR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84LTQQLTTPEE209 pKa = 4.51GQPDD213 pKa = 3.92TNPAIGEE220 pKa = 4.27QATGPAPILSPNPALGEE237 pKa = 4.09EE238 pKa = 4.29KK239 pKa = 10.76LEE241 pKa = 4.73SIQRR245 pKa = 11.84WLSFSPSDD253 pKa = 3.93SEE255 pKa = 4.3QDD257 pKa = 2.97IPAPPDD263 pKa = 3.4PEE265 pKa = 5.37RR266 pKa = 11.84PTLPQEE272 pKa = 4.45VFRR275 pKa = 11.84VPPSNSIPSVIAPSLAGGSLRR296 pKa = 11.84GSLFGGSLKK305 pKa = 10.3PRR307 pKa = 11.84SLDD310 pKa = 3.39SGKK313 pKa = 10.58LPEE316 pKa = 4.21TTPRR320 pKa = 11.84DD321 pKa = 3.82NVSVAGSLTGSLVGGSLRR339 pKa = 11.84GGALKK344 pKa = 10.54SPRR347 pKa = 11.84LDD349 pKa = 2.97TAQAKK354 pKa = 8.46EE355 pKa = 3.75HH356 pKa = 6.35ARR358 pKa = 11.84ILKK361 pKa = 9.93QLGKK365 pKa = 8.1TAAAEE370 pKa = 3.8YY371 pKa = 10.15LEE373 pKa = 4.43RR374 pKa = 11.84VRR376 pKa = 11.84ASQSSGQDD384 pKa = 3.09FKK386 pKa = 11.88
Molecular weight: 42.19 kDa
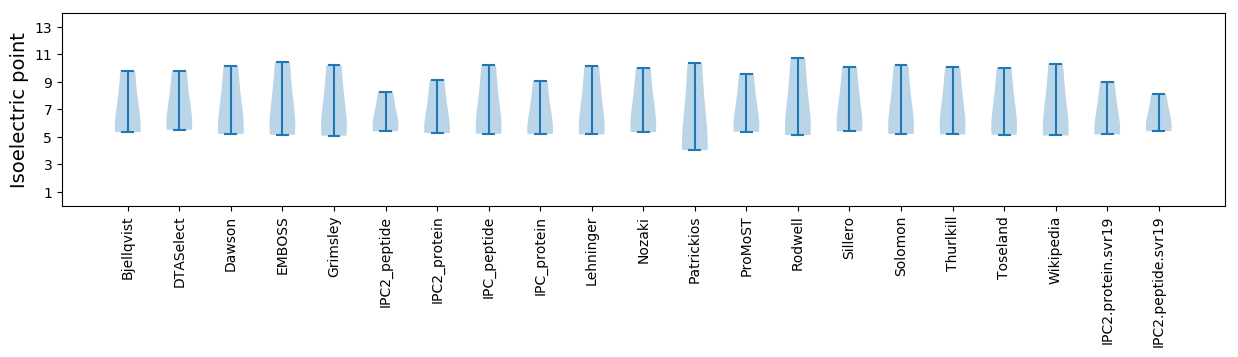
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEC6|A0A1L3KEC6_9VIRU Uncharacterized protein OS=Changjiang polero-like virus 1 OX=1922799 PE=4 SV=1
MM1 pKa = 7.34VIAISWMFGGLPGFLFGVALWLLKK25 pKa = 10.77SGWRR29 pKa = 11.84VLTSGPSYY37 pKa = 10.1MKK39 pKa = 9.8EE40 pKa = 3.84KK41 pKa = 10.69AVAGYY46 pKa = 10.38LSYY49 pKa = 10.4TIPQEE54 pKa = 4.03PPKK57 pKa = 10.57KK58 pKa = 10.5SIVLIQNSDD67 pKa = 3.35GSHH70 pKa = 6.46AGYY73 pKa = 7.72GTCVRR78 pKa = 11.84LYY80 pKa = 10.64NGEE83 pKa = 4.26TALLTADD90 pKa = 3.91HH91 pKa = 6.77VYY93 pKa = 10.96DD94 pKa = 3.93PALLVVSARR103 pKa = 11.84NGTKK107 pKa = 10.07VPLSQFQPVVRR118 pKa = 11.84GKK120 pKa = 8.78EE121 pKa = 3.57TDD123 pKa = 4.22LIILRR128 pKa = 11.84GPPNWEE134 pKa = 3.49GLLATKK140 pKa = 10.14AVQTVPVSQLAKK152 pKa = 10.45SKK154 pKa = 10.51FSLYY158 pKa = 10.83SFDD161 pKa = 4.95GAWQATNGEE170 pKa = 4.35LVGVEE175 pKa = 4.12GGFATTLCNTTPGFSGTPLFSGKK198 pKa = 9.73RR199 pKa = 11.84VVGVHH204 pKa = 5.67VGASKK209 pKa = 10.47KK210 pKa = 11.03ANVNLMAPIPSIAGLTSPQYY230 pKa = 10.93VFEE233 pKa = 4.4TTAPTGRR240 pKa = 11.84LFTSDD245 pKa = 3.25EE246 pKa = 4.06AVQLSEE252 pKa = 4.26TFQVLHH258 pKa = 6.61SEE260 pKa = 4.29LEE262 pKa = 3.97KK263 pKa = 11.17VLNFKK268 pKa = 10.29SQSGKK273 pKa = 8.72QWSEE277 pKa = 3.58MLDD280 pKa = 3.19IEE282 pKa = 4.66TTPTAPTPAVKK293 pKa = 10.47APVPAIQGNEE303 pKa = 3.55PRR305 pKa = 11.84GADD308 pKa = 2.87RR309 pKa = 11.84VINGEE314 pKa = 3.91ATVDD318 pKa = 3.49SGSTVATRR326 pKa = 11.84RR327 pKa = 11.84TSPVDD332 pKa = 3.43GEE334 pKa = 5.12DD335 pKa = 3.24ILAKK339 pKa = 10.75LMASLMSKK347 pKa = 10.39VDD349 pKa = 3.3VKK351 pKa = 11.08SIEE354 pKa = 3.99KK355 pKa = 9.88EE356 pKa = 3.95AIRR359 pKa = 11.84MISEE363 pKa = 4.32KK364 pKa = 10.62ALKK367 pKa = 9.88APRR370 pKa = 11.84RR371 pKa = 11.84RR372 pKa = 11.84GPRR375 pKa = 11.84KK376 pKa = 9.16PRR378 pKa = 11.84TSRR381 pKa = 11.84PSSIPTTPGKK391 pKa = 10.16YY392 pKa = 9.31VPPQRR397 pKa = 11.84RR398 pKa = 11.84VTHH401 pKa = 5.32QQASSGPANSPASTTLNKK419 pKa = 9.68EE420 pKa = 4.19AKK422 pKa = 9.73QSGGKK427 pKa = 9.43HH428 pKa = 5.06SPGTIQRR435 pKa = 11.84WVRR438 pKa = 11.84KK439 pKa = 9.54SEE441 pKa = 3.85ASAGRR446 pKa = 11.84CSAPKK451 pKa = 10.24QSS453 pKa = 3.47
MM1 pKa = 7.34VIAISWMFGGLPGFLFGVALWLLKK25 pKa = 10.77SGWRR29 pKa = 11.84VLTSGPSYY37 pKa = 10.1MKK39 pKa = 9.8EE40 pKa = 3.84KK41 pKa = 10.69AVAGYY46 pKa = 10.38LSYY49 pKa = 10.4TIPQEE54 pKa = 4.03PPKK57 pKa = 10.57KK58 pKa = 10.5SIVLIQNSDD67 pKa = 3.35GSHH70 pKa = 6.46AGYY73 pKa = 7.72GTCVRR78 pKa = 11.84LYY80 pKa = 10.64NGEE83 pKa = 4.26TALLTADD90 pKa = 3.91HH91 pKa = 6.77VYY93 pKa = 10.96DD94 pKa = 3.93PALLVVSARR103 pKa = 11.84NGTKK107 pKa = 10.07VPLSQFQPVVRR118 pKa = 11.84GKK120 pKa = 8.78EE121 pKa = 3.57TDD123 pKa = 4.22LIILRR128 pKa = 11.84GPPNWEE134 pKa = 3.49GLLATKK140 pKa = 10.14AVQTVPVSQLAKK152 pKa = 10.45SKK154 pKa = 10.51FSLYY158 pKa = 10.83SFDD161 pKa = 4.95GAWQATNGEE170 pKa = 4.35LVGVEE175 pKa = 4.12GGFATTLCNTTPGFSGTPLFSGKK198 pKa = 9.73RR199 pKa = 11.84VVGVHH204 pKa = 5.67VGASKK209 pKa = 10.47KK210 pKa = 11.03ANVNLMAPIPSIAGLTSPQYY230 pKa = 10.93VFEE233 pKa = 4.4TTAPTGRR240 pKa = 11.84LFTSDD245 pKa = 3.25EE246 pKa = 4.06AVQLSEE252 pKa = 4.26TFQVLHH258 pKa = 6.61SEE260 pKa = 4.29LEE262 pKa = 3.97KK263 pKa = 11.17VLNFKK268 pKa = 10.29SQSGKK273 pKa = 8.72QWSEE277 pKa = 3.58MLDD280 pKa = 3.19IEE282 pKa = 4.66TTPTAPTPAVKK293 pKa = 10.47APVPAIQGNEE303 pKa = 3.55PRR305 pKa = 11.84GADD308 pKa = 2.87RR309 pKa = 11.84VINGEE314 pKa = 3.91ATVDD318 pKa = 3.49SGSTVATRR326 pKa = 11.84RR327 pKa = 11.84TSPVDD332 pKa = 3.43GEE334 pKa = 5.12DD335 pKa = 3.24ILAKK339 pKa = 10.75LMASLMSKK347 pKa = 10.39VDD349 pKa = 3.3VKK351 pKa = 11.08SIEE354 pKa = 3.99KK355 pKa = 9.88EE356 pKa = 3.95AIRR359 pKa = 11.84MISEE363 pKa = 4.32KK364 pKa = 10.62ALKK367 pKa = 9.88APRR370 pKa = 11.84RR371 pKa = 11.84RR372 pKa = 11.84GPRR375 pKa = 11.84KK376 pKa = 9.16PRR378 pKa = 11.84TSRR381 pKa = 11.84PSSIPTTPGKK391 pKa = 10.16YY392 pKa = 9.31VPPQRR397 pKa = 11.84RR398 pKa = 11.84VTHH401 pKa = 5.32QQASSGPANSPASTTLNKK419 pKa = 9.68EE420 pKa = 4.19AKK422 pKa = 9.73QSGGKK427 pKa = 9.43HH428 pKa = 5.06SPGTIQRR435 pKa = 11.84WVRR438 pKa = 11.84KK439 pKa = 9.54SEE441 pKa = 3.85ASAGRR446 pKa = 11.84CSAPKK451 pKa = 10.24QSS453 pKa = 3.47
Molecular weight: 48.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1220 |
381 |
453 |
406.7 |
44.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.295 ± 0.686 | 1.393 ± 0.325 |
4.754 ± 0.94 | 5.656 ± 0.363 |
3.279 ± 0.255 | 7.787 ± 0.758 |
1.967 ± 0.636 | 3.934 ± 0.438 |
5.246 ± 0.824 | 9.344 ± 1.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.885 ± 0.182 | 3.607 ± 0.335 |
7.049 ± 0.768 | 4.18 ± 0.124 |
5.164 ± 0.242 | 10.082 ± 0.535 |
5.656 ± 1.031 | 7.295 ± 0.889 |
1.967 ± 0.198 | 2.459 ± 0.263 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
