
Abyssibacter profundi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Salinisphaerales; Salinisphaeraceae; Abyssibacter
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
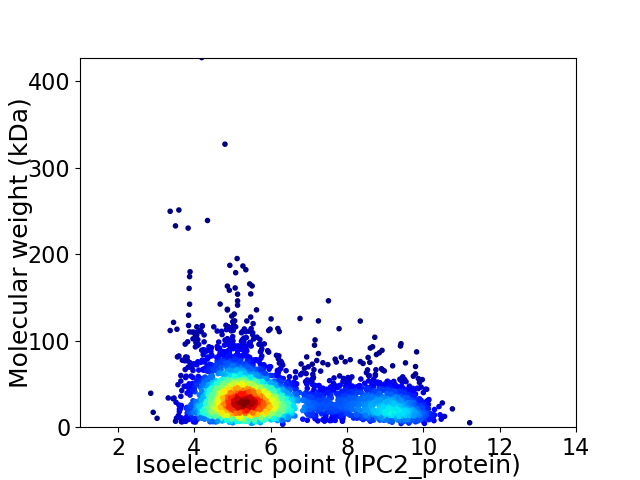
Virtual 2D-PAGE plot for 3331 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A383XPS3|A0A383XPS3_9GAMM Uncharacterized protein OS=Abyssibacter profundi OX=2182787 GN=DEH80_16535 PE=4 SV=1
MM1 pKa = 7.3FEE3 pKa = 4.18SARR6 pKa = 11.84SRR8 pKa = 11.84VLIAGIASTCSLSALAQQSGPTACDD33 pKa = 2.84IAFGAVEE40 pKa = 4.3SGAVAAAIDD49 pKa = 4.01PALQPGKK56 pKa = 9.08FQRR59 pKa = 11.84IVLTFADD66 pKa = 3.82AAARR70 pKa = 11.84DD71 pKa = 3.66AALEE75 pKa = 4.0RR76 pKa = 11.84VRR78 pKa = 11.84LANFLPVDD86 pKa = 4.03VARR89 pKa = 11.84DD90 pKa = 3.83AYY92 pKa = 10.78AMRR95 pKa = 11.84HH96 pKa = 5.25LNFVVLPALTVSTDD110 pKa = 4.29LLDD113 pKa = 3.48QLAALGRR120 pKa = 11.84DD121 pKa = 3.26LGLISVSTDD130 pKa = 3.05PEE132 pKa = 4.14VRR134 pKa = 11.84SLIDD138 pKa = 3.04STGPLIGVNAARR150 pKa = 11.84AAFGEE155 pKa = 4.46GKK157 pKa = 10.06EE158 pKa = 4.02PAGYY162 pKa = 7.95TVTGDD167 pKa = 3.9GIGVAVLDD175 pKa = 4.13TGWDD179 pKa = 3.24KK180 pKa = 10.16TQGDD184 pKa = 4.51FEE186 pKa = 4.5HH187 pKa = 7.19LEE189 pKa = 4.1TFGNVRR195 pKa = 11.84MVGPQAIEE203 pKa = 3.63FVPPVEE209 pKa = 4.5NSEE212 pKa = 4.31TNQGHH217 pKa = 5.43GTHH220 pKa = 6.31IVGTIAGNGAQSDD233 pKa = 3.86GRR235 pKa = 11.84IVGLAPQVNMLSVAVDD251 pKa = 3.74AGASYY256 pKa = 11.58AFLLDD261 pKa = 3.4GFDD264 pKa = 3.97YY265 pKa = 11.07VLEE268 pKa = 4.32VQDD271 pKa = 4.82DD272 pKa = 3.98YY273 pKa = 11.88NIRR276 pKa = 11.84VTNHH280 pKa = 6.04SYY282 pKa = 11.19GPAVGSAFRR291 pKa = 11.84FDD293 pKa = 4.1PSTASSQAIRR303 pKa = 11.84AFYY306 pKa = 9.37QAGIIPVFAAGNAGPDD322 pKa = 3.71EE323 pKa = 4.4DD324 pKa = 5.29TISADD329 pKa = 3.39AQNPCAIGVAAGTRR343 pKa = 11.84ALTLTDD349 pKa = 4.42FSSRR353 pKa = 11.84GRR355 pKa = 11.84SDD357 pKa = 3.21NAIAGPDD364 pKa = 2.94ITAPGDD370 pKa = 3.91AITASRR376 pKa = 11.84AINGFTSTSIPDD388 pKa = 4.03LDD390 pKa = 3.83NPAYY394 pKa = 9.53ATISGTSMAAPHH406 pKa = 5.74VVASIALILEE416 pKa = 4.41ADD418 pKa = 3.63EE419 pKa = 4.58SLSFEE424 pKa = 4.6DD425 pKa = 4.14VYY427 pKa = 11.71EE428 pKa = 5.29LITSTATPMKK438 pKa = 10.37RR439 pKa = 11.84ADD441 pKa = 3.36QTAYY445 pKa = 10.26EE446 pKa = 4.09PFEE449 pKa = 4.25VGFGYY454 pKa = 10.1IDD456 pKa = 3.36TAAAIAKK463 pKa = 9.87LLGEE467 pKa = 4.29QKK469 pKa = 10.33PEE471 pKa = 3.52RR472 pKa = 11.84LQPVEE477 pKa = 4.47CPAPGEE483 pKa = 4.06SSVINFAATAAAVYY497 pKa = 7.41STPAVSFSPSGFEE510 pKa = 3.78DD511 pKa = 3.62SIYY514 pKa = 10.91QFLLEE519 pKa = 4.63PSCALASMTVEE530 pKa = 4.41IEE532 pKa = 3.81WSATLPEE539 pKa = 5.75DD540 pKa = 4.85IDD542 pKa = 4.37LEE544 pKa = 4.71LIGPEE549 pKa = 4.09GGVIGASTNRR559 pKa = 11.84QLEE562 pKa = 4.24EE563 pKa = 3.76GAEE566 pKa = 3.98PSEE569 pKa = 4.0AVTIEE574 pKa = 4.11SAGGGLYY581 pKa = 9.53TSRR584 pKa = 11.84VFGYY588 pKa = 10.51RR589 pKa = 11.84NNNLPYY595 pKa = 9.02TGTVTLVAEE604 pKa = 4.42QNINTAPVAALSVPATAGVDD624 pKa = 3.54EE625 pKa = 5.71EE626 pKa = 4.67ITVSAAASSDD636 pKa = 3.59PDD638 pKa = 3.72GDD640 pKa = 3.81ALNYY644 pKa = 10.37QFDD647 pKa = 4.57LGDD650 pKa = 3.78GTQTTVSTSPTVTHH664 pKa = 7.34RR665 pKa = 11.84YY666 pKa = 7.28ATAGDD671 pKa = 3.88YY672 pKa = 10.93VVTVTVSDD680 pKa = 3.55ASGATDD686 pKa = 3.73SASQTITVAEE696 pKa = 4.47DD697 pKa = 3.35GGGNTAGPGSTIDD710 pKa = 3.32AVLAVSGDD718 pKa = 3.62GAEE721 pKa = 4.24SDD723 pKa = 3.63GDD725 pKa = 4.1AVNGDD730 pKa = 3.75AEE732 pKa = 4.76TVFTFDD738 pKa = 5.08ASDD741 pKa = 3.08SGYY744 pKa = 9.34TDD746 pKa = 4.58AEE748 pKa = 4.41GSDD751 pKa = 3.72LYY753 pKa = 10.52YY754 pKa = 10.78TFVFGDD760 pKa = 3.56EE761 pKa = 4.21ASEE764 pKa = 4.2EE765 pKa = 4.69EE766 pKa = 4.59YY767 pKa = 11.12ASTTSDD773 pKa = 4.15PIATHH778 pKa = 6.63SYY780 pKa = 9.35GAAGTYY786 pKa = 7.26EE787 pKa = 4.3AYY789 pKa = 10.8VIVSDD794 pKa = 4.82AFGNSDD800 pKa = 3.39TSNTVTITTTITITVEE816 pKa = 3.83GDD818 pKa = 2.93NGTVAQLTVDD828 pKa = 4.03EE829 pKa = 4.76TSGTAPLQVVFDD841 pKa = 4.49GSQSFTSEE849 pKa = 4.07GEE851 pKa = 4.36TITEE855 pKa = 4.07YY856 pKa = 11.19CFDD859 pKa = 4.58FDD861 pKa = 5.92DD862 pKa = 6.4GEE864 pKa = 4.53APQCGTEE871 pKa = 4.01DD872 pKa = 3.45TATYY876 pKa = 10.56VYY878 pKa = 9.26TRR880 pKa = 11.84PGSYY884 pKa = 9.97EE885 pKa = 3.58PSLTVTASDD894 pKa = 4.06EE895 pKa = 4.31STATAKK901 pKa = 9.97ATVSVGGTGSNPGTPSQPAASTGGGSGALGGLLLLPLL938 pKa = 4.51
MM1 pKa = 7.3FEE3 pKa = 4.18SARR6 pKa = 11.84SRR8 pKa = 11.84VLIAGIASTCSLSALAQQSGPTACDD33 pKa = 2.84IAFGAVEE40 pKa = 4.3SGAVAAAIDD49 pKa = 4.01PALQPGKK56 pKa = 9.08FQRR59 pKa = 11.84IVLTFADD66 pKa = 3.82AAARR70 pKa = 11.84DD71 pKa = 3.66AALEE75 pKa = 4.0RR76 pKa = 11.84VRR78 pKa = 11.84LANFLPVDD86 pKa = 4.03VARR89 pKa = 11.84DD90 pKa = 3.83AYY92 pKa = 10.78AMRR95 pKa = 11.84HH96 pKa = 5.25LNFVVLPALTVSTDD110 pKa = 4.29LLDD113 pKa = 3.48QLAALGRR120 pKa = 11.84DD121 pKa = 3.26LGLISVSTDD130 pKa = 3.05PEE132 pKa = 4.14VRR134 pKa = 11.84SLIDD138 pKa = 3.04STGPLIGVNAARR150 pKa = 11.84AAFGEE155 pKa = 4.46GKK157 pKa = 10.06EE158 pKa = 4.02PAGYY162 pKa = 7.95TVTGDD167 pKa = 3.9GIGVAVLDD175 pKa = 4.13TGWDD179 pKa = 3.24KK180 pKa = 10.16TQGDD184 pKa = 4.51FEE186 pKa = 4.5HH187 pKa = 7.19LEE189 pKa = 4.1TFGNVRR195 pKa = 11.84MVGPQAIEE203 pKa = 3.63FVPPVEE209 pKa = 4.5NSEE212 pKa = 4.31TNQGHH217 pKa = 5.43GTHH220 pKa = 6.31IVGTIAGNGAQSDD233 pKa = 3.86GRR235 pKa = 11.84IVGLAPQVNMLSVAVDD251 pKa = 3.74AGASYY256 pKa = 11.58AFLLDD261 pKa = 3.4GFDD264 pKa = 3.97YY265 pKa = 11.07VLEE268 pKa = 4.32VQDD271 pKa = 4.82DD272 pKa = 3.98YY273 pKa = 11.88NIRR276 pKa = 11.84VTNHH280 pKa = 6.04SYY282 pKa = 11.19GPAVGSAFRR291 pKa = 11.84FDD293 pKa = 4.1PSTASSQAIRR303 pKa = 11.84AFYY306 pKa = 9.37QAGIIPVFAAGNAGPDD322 pKa = 3.71EE323 pKa = 4.4DD324 pKa = 5.29TISADD329 pKa = 3.39AQNPCAIGVAAGTRR343 pKa = 11.84ALTLTDD349 pKa = 4.42FSSRR353 pKa = 11.84GRR355 pKa = 11.84SDD357 pKa = 3.21NAIAGPDD364 pKa = 2.94ITAPGDD370 pKa = 3.91AITASRR376 pKa = 11.84AINGFTSTSIPDD388 pKa = 4.03LDD390 pKa = 3.83NPAYY394 pKa = 9.53ATISGTSMAAPHH406 pKa = 5.74VVASIALILEE416 pKa = 4.41ADD418 pKa = 3.63EE419 pKa = 4.58SLSFEE424 pKa = 4.6DD425 pKa = 4.14VYY427 pKa = 11.71EE428 pKa = 5.29LITSTATPMKK438 pKa = 10.37RR439 pKa = 11.84ADD441 pKa = 3.36QTAYY445 pKa = 10.26EE446 pKa = 4.09PFEE449 pKa = 4.25VGFGYY454 pKa = 10.1IDD456 pKa = 3.36TAAAIAKK463 pKa = 9.87LLGEE467 pKa = 4.29QKK469 pKa = 10.33PEE471 pKa = 3.52RR472 pKa = 11.84LQPVEE477 pKa = 4.47CPAPGEE483 pKa = 4.06SSVINFAATAAAVYY497 pKa = 7.41STPAVSFSPSGFEE510 pKa = 3.78DD511 pKa = 3.62SIYY514 pKa = 10.91QFLLEE519 pKa = 4.63PSCALASMTVEE530 pKa = 4.41IEE532 pKa = 3.81WSATLPEE539 pKa = 5.75DD540 pKa = 4.85IDD542 pKa = 4.37LEE544 pKa = 4.71LIGPEE549 pKa = 4.09GGVIGASTNRR559 pKa = 11.84QLEE562 pKa = 4.24EE563 pKa = 3.76GAEE566 pKa = 3.98PSEE569 pKa = 4.0AVTIEE574 pKa = 4.11SAGGGLYY581 pKa = 9.53TSRR584 pKa = 11.84VFGYY588 pKa = 10.51RR589 pKa = 11.84NNNLPYY595 pKa = 9.02TGTVTLVAEE604 pKa = 4.42QNINTAPVAALSVPATAGVDD624 pKa = 3.54EE625 pKa = 5.71EE626 pKa = 4.67ITVSAAASSDD636 pKa = 3.59PDD638 pKa = 3.72GDD640 pKa = 3.81ALNYY644 pKa = 10.37QFDD647 pKa = 4.57LGDD650 pKa = 3.78GTQTTVSTSPTVTHH664 pKa = 7.34RR665 pKa = 11.84YY666 pKa = 7.28ATAGDD671 pKa = 3.88YY672 pKa = 10.93VVTVTVSDD680 pKa = 3.55ASGATDD686 pKa = 3.73SASQTITVAEE696 pKa = 4.47DD697 pKa = 3.35GGGNTAGPGSTIDD710 pKa = 3.32AVLAVSGDD718 pKa = 3.62GAEE721 pKa = 4.24SDD723 pKa = 3.63GDD725 pKa = 4.1AVNGDD730 pKa = 3.75AEE732 pKa = 4.76TVFTFDD738 pKa = 5.08ASDD741 pKa = 3.08SGYY744 pKa = 9.34TDD746 pKa = 4.58AEE748 pKa = 4.41GSDD751 pKa = 3.72LYY753 pKa = 10.52YY754 pKa = 10.78TFVFGDD760 pKa = 3.56EE761 pKa = 4.21ASEE764 pKa = 4.2EE765 pKa = 4.69EE766 pKa = 4.59YY767 pKa = 11.12ASTTSDD773 pKa = 4.15PIATHH778 pKa = 6.63SYY780 pKa = 9.35GAAGTYY786 pKa = 7.26EE787 pKa = 4.3AYY789 pKa = 10.8VIVSDD794 pKa = 4.82AFGNSDD800 pKa = 3.39TSNTVTITTTITITVEE816 pKa = 3.83GDD818 pKa = 2.93NGTVAQLTVDD828 pKa = 4.03EE829 pKa = 4.76TSGTAPLQVVFDD841 pKa = 4.49GSQSFTSEE849 pKa = 4.07GEE851 pKa = 4.36TITEE855 pKa = 4.07YY856 pKa = 11.19CFDD859 pKa = 4.58FDD861 pKa = 5.92DD862 pKa = 6.4GEE864 pKa = 4.53APQCGTEE871 pKa = 4.01DD872 pKa = 3.45TATYY876 pKa = 10.56VYY878 pKa = 9.26TRR880 pKa = 11.84PGSYY884 pKa = 9.97EE885 pKa = 3.58PSLTVTASDD894 pKa = 4.06EE895 pKa = 4.31STATAKK901 pKa = 9.97ATVSVGGTGSNPGTPSQPAASTGGGSGALGGLLLLPLL938 pKa = 4.51
Molecular weight: 96.1 kDa
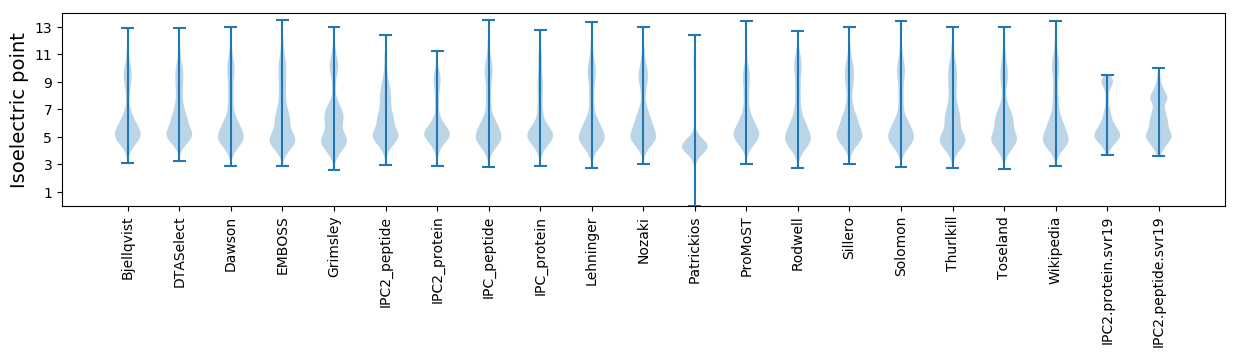
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A383XR84|A0A383XR84_9GAMM TetR/AcrR family transcriptional regulator OS=Abyssibacter profundi OX=2182787 GN=DEH80_14020 PE=4 SV=1
MM1 pKa = 7.26KK2 pKa = 9.53QTFQPHH8 pKa = 5.02AVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.53RR14 pKa = 11.84THH16 pKa = 5.8GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.18VIKK32 pKa = 10.13ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.81GRR39 pKa = 11.84KK40 pKa = 9.07RR41 pKa = 11.84LTPP44 pKa = 3.95
MM1 pKa = 7.26KK2 pKa = 9.53QTFQPHH8 pKa = 5.02AVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.53RR14 pKa = 11.84THH16 pKa = 5.8GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.18VIKK32 pKa = 10.13ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.81GRR39 pKa = 11.84KK40 pKa = 9.07RR41 pKa = 11.84LTPP44 pKa = 3.95
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1138768 |
33 |
4044 |
341.9 |
37.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.074 ± 0.058 | 0.948 ± 0.014 |
6.336 ± 0.046 | 5.723 ± 0.039 |
3.308 ± 0.024 | 8.296 ± 0.047 |
2.184 ± 0.021 | 4.432 ± 0.03 |
2.252 ± 0.033 | 10.579 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.216 ± 0.018 | 2.509 ± 0.023 |
5.263 ± 0.036 | 4.214 ± 0.03 |
7.429 ± 0.045 | 5.528 ± 0.033 |
5.425 ± 0.04 | 7.503 ± 0.034 |
1.458 ± 0.019 | 2.324 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
