
Rotavirus A turkey-tc/GER/03V0002E10/2003/G22P[35]
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus A; Rotavirus A isolates
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
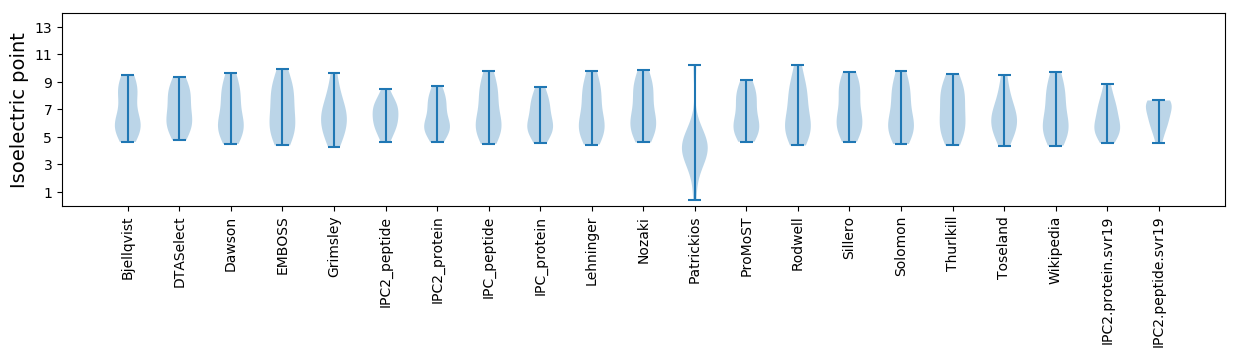
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|L0CQJ0|L0CQJ0_9REOV Non-structural protein 5 OS=Rotavirus A turkey-tc/GER/03V0002E10/2003/G22P[35] OX=1271535 PE=4 SV=1
MM1 pKa = 7.48YY2 pKa = 9.47STEE5 pKa = 4.05CTILSIEE12 pKa = 4.27IIFYY16 pKa = 10.62CLAAILLYY24 pKa = 11.28DD25 pKa = 3.93MLCRR29 pKa = 11.84MANSPLFCFSVLIGVVTILPRR50 pKa = 11.84CYY52 pKa = 10.14AQNYY56 pKa = 7.88GINVPITGSLDD67 pKa = 3.25VTVQNQTSEE76 pKa = 4.72PIGLTSTLCIYY87 pKa = 10.87YY88 pKa = 8.33PTEE91 pKa = 4.02AANEE95 pKa = 4.11IADD98 pKa = 4.27SEE100 pKa = 4.43WKK102 pKa = 8.15QTVTQLFLTKK112 pKa = 9.98GWPTTSIYY120 pKa = 10.86LNEE123 pKa = 4.18YY124 pKa = 9.69TDD126 pKa = 4.62LQTFSNNPQLNCDD139 pKa = 3.73YY140 pKa = 11.12NIVLVKK146 pKa = 10.63YY147 pKa = 10.35DD148 pKa = 3.86GNQGLDD154 pKa = 2.95ISEE157 pKa = 4.24LAEE160 pKa = 3.99LLLYY164 pKa = 10.19EE165 pKa = 4.32WLCNEE170 pKa = 4.36MDD172 pKa = 3.29VSLYY176 pKa = 10.67YY177 pKa = 9.88YY178 pKa = 10.06QQVSEE183 pKa = 4.16ANKK186 pKa = 8.42WIAMGTDD193 pKa = 3.77CTIKK197 pKa = 10.42VCPLNTQTLGIGCKK211 pKa = 7.88TTDD214 pKa = 3.2LTTFEE219 pKa = 4.48QLTASEE225 pKa = 4.47KK226 pKa = 10.41LAIVDD231 pKa = 3.63VVDD234 pKa = 4.5GVNHH238 pKa = 6.81KK239 pKa = 9.93IDD241 pKa = 3.84YY242 pKa = 8.11TVTTCNVKK250 pKa = 10.05NCLRR254 pKa = 11.84LSQRR258 pKa = 11.84EE259 pKa = 4.01NVAIIQVGGPEE270 pKa = 4.31VIDD273 pKa = 3.73VSEE276 pKa = 5.04DD277 pKa = 3.34PMVVPKK283 pKa = 8.42MQRR286 pKa = 11.84VTRR289 pKa = 11.84INWKK293 pKa = 9.4KK294 pKa = 7.32WWQVFYY300 pKa = 10.64TIVDD304 pKa = 3.85YY305 pKa = 11.16INTIIQTMSKK315 pKa = 10.31RR316 pKa = 11.84SRR318 pKa = 11.84SLNASAYY325 pKa = 8.0YY326 pKa = 10.36FRR328 pKa = 11.84VV329 pKa = 3.31
MM1 pKa = 7.48YY2 pKa = 9.47STEE5 pKa = 4.05CTILSIEE12 pKa = 4.27IIFYY16 pKa = 10.62CLAAILLYY24 pKa = 11.28DD25 pKa = 3.93MLCRR29 pKa = 11.84MANSPLFCFSVLIGVVTILPRR50 pKa = 11.84CYY52 pKa = 10.14AQNYY56 pKa = 7.88GINVPITGSLDD67 pKa = 3.25VTVQNQTSEE76 pKa = 4.72PIGLTSTLCIYY87 pKa = 10.87YY88 pKa = 8.33PTEE91 pKa = 4.02AANEE95 pKa = 4.11IADD98 pKa = 4.27SEE100 pKa = 4.43WKK102 pKa = 8.15QTVTQLFLTKK112 pKa = 9.98GWPTTSIYY120 pKa = 10.86LNEE123 pKa = 4.18YY124 pKa = 9.69TDD126 pKa = 4.62LQTFSNNPQLNCDD139 pKa = 3.73YY140 pKa = 11.12NIVLVKK146 pKa = 10.63YY147 pKa = 10.35DD148 pKa = 3.86GNQGLDD154 pKa = 2.95ISEE157 pKa = 4.24LAEE160 pKa = 3.99LLLYY164 pKa = 10.19EE165 pKa = 4.32WLCNEE170 pKa = 4.36MDD172 pKa = 3.29VSLYY176 pKa = 10.67YY177 pKa = 9.88YY178 pKa = 10.06QQVSEE183 pKa = 4.16ANKK186 pKa = 8.42WIAMGTDD193 pKa = 3.77CTIKK197 pKa = 10.42VCPLNTQTLGIGCKK211 pKa = 7.88TTDD214 pKa = 3.2LTTFEE219 pKa = 4.48QLTASEE225 pKa = 4.47KK226 pKa = 10.41LAIVDD231 pKa = 3.63VVDD234 pKa = 4.5GVNHH238 pKa = 6.81KK239 pKa = 9.93IDD241 pKa = 3.84YY242 pKa = 8.11TVTTCNVKK250 pKa = 10.05NCLRR254 pKa = 11.84LSQRR258 pKa = 11.84EE259 pKa = 4.01NVAIIQVGGPEE270 pKa = 4.31VIDD273 pKa = 3.73VSEE276 pKa = 5.04DD277 pKa = 3.34PMVVPKK283 pKa = 8.42MQRR286 pKa = 11.84VTRR289 pKa = 11.84INWKK293 pKa = 9.4KK294 pKa = 7.32WWQVFYY300 pKa = 10.64TIVDD304 pKa = 3.85YY305 pKa = 11.16INTIIQTMSKK315 pKa = 10.31RR316 pKa = 11.84SRR318 pKa = 11.84SLNASAYY325 pKa = 8.0YY326 pKa = 10.36FRR328 pKa = 11.84VV329 pKa = 3.31
Molecular weight: 37.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0CS97|L0CS97_9REOV Intermediate capsid protein VP6 OS=Rotavirus A turkey-tc/GER/03V0002E10/2003/G22P[35] OX=1271535 PE=3 SV=1
MM1 pKa = 6.85NHH3 pKa = 5.71LVKK6 pKa = 10.64RR7 pKa = 11.84QVLQEE12 pKa = 3.9NLLVGVSSMYY22 pKa = 10.39HH23 pKa = 5.96CPKK26 pKa = 9.77KK27 pKa = 10.27HH28 pKa = 6.21LVNTCLIGIQKK39 pKa = 8.46TLDD42 pKa = 3.36HH43 pKa = 7.23LIQHH47 pKa = 6.18QMTCPPNLLLNQMQLNQMLMLVCRR71 pKa = 11.84WMLVQQIVRR80 pKa = 11.84QEE82 pKa = 3.77AMQKK86 pKa = 8.72MKK88 pKa = 10.82SIFHH92 pKa = 5.94SLKK95 pKa = 10.27EE96 pKa = 3.95
MM1 pKa = 6.85NHH3 pKa = 5.71LVKK6 pKa = 10.64RR7 pKa = 11.84QVLQEE12 pKa = 3.9NLLVGVSSMYY22 pKa = 10.39HH23 pKa = 5.96CPKK26 pKa = 9.77KK27 pKa = 10.27HH28 pKa = 6.21LVNTCLIGIQKK39 pKa = 8.46TLDD42 pKa = 3.36HH43 pKa = 7.23LIQHH47 pKa = 6.18QMTCPPNLLLNQMQLNQMLMLVCRR71 pKa = 11.84WMLVQQIVRR80 pKa = 11.84QEE82 pKa = 3.77AMQKK86 pKa = 8.72MKK88 pKa = 10.82SIFHH92 pKa = 5.94SLKK95 pKa = 10.27EE96 pKa = 3.95
Molecular weight: 11.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5987 |
96 |
1089 |
498.9 |
57.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.428 ± 0.318 | 1.353 ± 0.424 |
6.314 ± 0.274 | 5.345 ± 0.238 |
4.092 ± 0.232 | 3.341 ± 0.391 |
1.57 ± 0.278 | 7.717 ± 0.221 |
6.731 ± 0.557 | 9.237 ± 0.37 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.739 ± 0.247 | 6.798 ± 0.339 |
3.207 ± 0.294 | 4.343 ± 0.491 |
4.76 ± 0.315 | 7.65 ± 0.737 |
6.347 ± 0.415 | 6.648 ± 0.346 |
1.086 ± 0.165 | 5.295 ± 0.688 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
