
Common bean-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; unclassified Gemycircularvirus
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
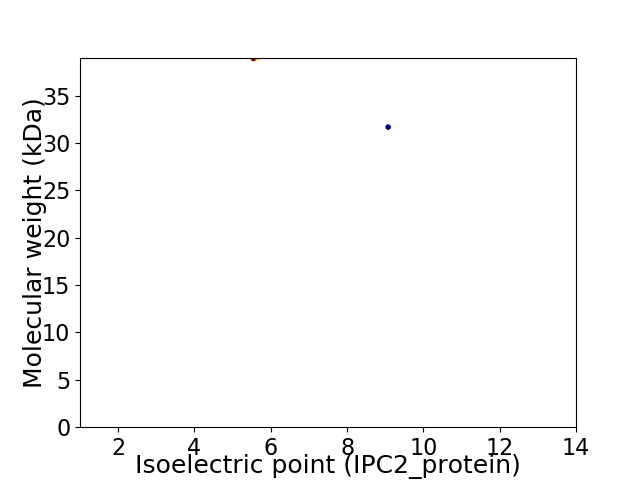
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
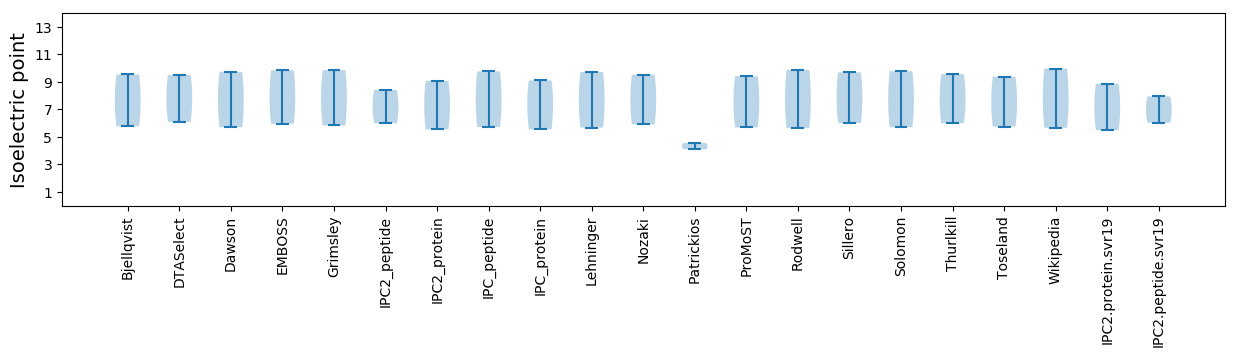
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C8YZ07|A0A1C8YZ07_9VIRU Capsid protein OS=Common bean-associated gemycircularvirus OX=1897538 PE=4 SV=1
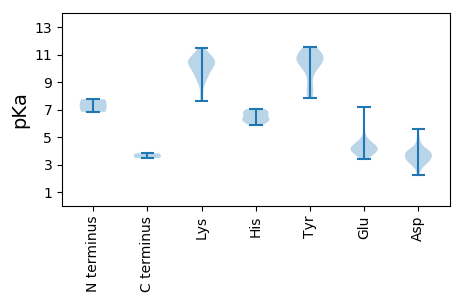
MM1 pKa = 6.83PAKK4 pKa = 10.54YY5 pKa = 9.99KK6 pKa = 11.17LNDD9 pKa = 3.34EE10 pKa = 4.28QFFMLTYY17 pKa = 8.23PTIPDD22 pKa = 3.68GFDD25 pKa = 3.07TDD27 pKa = 3.98GLINVLEE34 pKa = 4.13RR35 pKa = 11.84LGCNYY40 pKa = 10.45RR41 pKa = 11.84IGRR44 pKa = 11.84EE45 pKa = 3.79LHH47 pKa = 6.16QDD49 pKa = 3.95GKK51 pKa = 9.65PHH53 pKa = 6.18LHH55 pKa = 7.03AMLCFDD61 pKa = 4.92EE62 pKa = 5.88PYY64 pKa = 10.44TDD66 pKa = 3.55GDD68 pKa = 3.29ARR70 pKa = 11.84ATFKK74 pKa = 10.69IGTRR78 pKa = 11.84VPNIRR83 pKa = 11.84VRR85 pKa = 11.84RR86 pKa = 11.84VKK88 pKa = 10.46PEE90 pKa = 3.96RR91 pKa = 11.84GWDD94 pKa = 3.59YY95 pKa = 11.17VGKK98 pKa = 9.63HH99 pKa = 5.89AGTKK103 pKa = 8.06EE104 pKa = 3.4GHH106 pKa = 6.39YY107 pKa = 10.45IVGEE111 pKa = 3.92KK112 pKa = 9.01GTRR115 pKa = 11.84PGGDD119 pKa = 3.14EE120 pKa = 4.65DD121 pKa = 3.72STEE124 pKa = 4.08RR125 pKa = 11.84SSNDD129 pKa = 2.27AWFEE133 pKa = 4.08IILARR138 pKa = 11.84TRR140 pKa = 11.84EE141 pKa = 4.02EE142 pKa = 4.24FFDD145 pKa = 3.61SAARR149 pKa = 11.84LAPRR153 pKa = 11.84QLACNFSSLTAYY165 pKa = 10.43ADD167 pKa = 3.1WKK169 pKa = 9.37YY170 pKa = 11.01RR171 pKa = 11.84PVAAAYY177 pKa = 9.4SNPDD181 pKa = 3.11GEE183 pKa = 4.29FSVPFEE189 pKa = 4.33LKK191 pKa = 10.48DD192 pKa = 3.42WVDD195 pKa = 3.86DD196 pKa = 4.02NVRR199 pKa = 11.84GCSGRR204 pKa = 11.84PKK206 pKa = 10.77GLVLFGATRR215 pKa = 11.84LGKK218 pKa = 7.65TVWARR223 pKa = 11.84SLGNHH228 pKa = 6.22YY229 pKa = 10.94YY230 pKa = 10.72CGGLWNLADD239 pKa = 3.85FDD241 pKa = 4.31EE242 pKa = 4.69TVEE245 pKa = 3.99YY246 pKa = 10.68AIFDD250 pKa = 3.91DD251 pKa = 4.15MVNGLRR257 pKa = 11.84AGYY260 pKa = 8.54FQYY263 pKa = 11.18KK264 pKa = 9.87DD265 pKa = 3.33WLGGQYY271 pKa = 10.9EE272 pKa = 4.5FMVQDD277 pKa = 3.39KK278 pKa = 10.52YY279 pKa = 10.9KK280 pKa = 10.72GKK282 pKa = 10.48RR283 pKa = 11.84RR284 pKa = 11.84VKK286 pKa = 8.91WGKK289 pKa = 9.53PSIFICNRR297 pKa = 11.84DD298 pKa = 3.38PRR300 pKa = 11.84DD301 pKa = 3.66EE302 pKa = 5.03IGLDD306 pKa = 3.44DD307 pKa = 4.42HH308 pKa = 6.89NKK310 pKa = 10.55SKK312 pKa = 10.91IEE314 pKa = 3.86WDD316 pKa = 3.12WMEE319 pKa = 4.28EE320 pKa = 3.59NCLFYY325 pKa = 10.65EE326 pKa = 3.95LRR328 pKa = 11.84EE329 pKa = 4.24PIFRR333 pKa = 11.84ASTEE337 pKa = 3.81
MM1 pKa = 6.83PAKK4 pKa = 10.54YY5 pKa = 9.99KK6 pKa = 11.17LNDD9 pKa = 3.34EE10 pKa = 4.28QFFMLTYY17 pKa = 8.23PTIPDD22 pKa = 3.68GFDD25 pKa = 3.07TDD27 pKa = 3.98GLINVLEE34 pKa = 4.13RR35 pKa = 11.84LGCNYY40 pKa = 10.45RR41 pKa = 11.84IGRR44 pKa = 11.84EE45 pKa = 3.79LHH47 pKa = 6.16QDD49 pKa = 3.95GKK51 pKa = 9.65PHH53 pKa = 6.18LHH55 pKa = 7.03AMLCFDD61 pKa = 4.92EE62 pKa = 5.88PYY64 pKa = 10.44TDD66 pKa = 3.55GDD68 pKa = 3.29ARR70 pKa = 11.84ATFKK74 pKa = 10.69IGTRR78 pKa = 11.84VPNIRR83 pKa = 11.84VRR85 pKa = 11.84RR86 pKa = 11.84VKK88 pKa = 10.46PEE90 pKa = 3.96RR91 pKa = 11.84GWDD94 pKa = 3.59YY95 pKa = 11.17VGKK98 pKa = 9.63HH99 pKa = 5.89AGTKK103 pKa = 8.06EE104 pKa = 3.4GHH106 pKa = 6.39YY107 pKa = 10.45IVGEE111 pKa = 3.92KK112 pKa = 9.01GTRR115 pKa = 11.84PGGDD119 pKa = 3.14EE120 pKa = 4.65DD121 pKa = 3.72STEE124 pKa = 4.08RR125 pKa = 11.84SSNDD129 pKa = 2.27AWFEE133 pKa = 4.08IILARR138 pKa = 11.84TRR140 pKa = 11.84EE141 pKa = 4.02EE142 pKa = 4.24FFDD145 pKa = 3.61SAARR149 pKa = 11.84LAPRR153 pKa = 11.84QLACNFSSLTAYY165 pKa = 10.43ADD167 pKa = 3.1WKK169 pKa = 9.37YY170 pKa = 11.01RR171 pKa = 11.84PVAAAYY177 pKa = 9.4SNPDD181 pKa = 3.11GEE183 pKa = 4.29FSVPFEE189 pKa = 4.33LKK191 pKa = 10.48DD192 pKa = 3.42WVDD195 pKa = 3.86DD196 pKa = 4.02NVRR199 pKa = 11.84GCSGRR204 pKa = 11.84PKK206 pKa = 10.77GLVLFGATRR215 pKa = 11.84LGKK218 pKa = 7.65TVWARR223 pKa = 11.84SLGNHH228 pKa = 6.22YY229 pKa = 10.94YY230 pKa = 10.72CGGLWNLADD239 pKa = 3.85FDD241 pKa = 4.31EE242 pKa = 4.69TVEE245 pKa = 3.99YY246 pKa = 10.68AIFDD250 pKa = 3.91DD251 pKa = 4.15MVNGLRR257 pKa = 11.84AGYY260 pKa = 8.54FQYY263 pKa = 11.18KK264 pKa = 9.87DD265 pKa = 3.33WLGGQYY271 pKa = 10.9EE272 pKa = 4.5FMVQDD277 pKa = 3.39KK278 pKa = 10.52YY279 pKa = 10.9KK280 pKa = 10.72GKK282 pKa = 10.48RR283 pKa = 11.84RR284 pKa = 11.84VKK286 pKa = 8.91WGKK289 pKa = 9.53PSIFICNRR297 pKa = 11.84DD298 pKa = 3.38PRR300 pKa = 11.84DD301 pKa = 3.66EE302 pKa = 5.03IGLDD306 pKa = 3.44DD307 pKa = 4.42HH308 pKa = 6.89NKK310 pKa = 10.55SKK312 pKa = 10.91IEE314 pKa = 3.86WDD316 pKa = 3.12WMEE319 pKa = 4.28EE320 pKa = 3.59NCLFYY325 pKa = 10.65EE326 pKa = 3.95LRR328 pKa = 11.84EE329 pKa = 4.24PIFRR333 pKa = 11.84ASTEE337 pKa = 3.81
Molecular weight: 38.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C8YZ07|A0A1C8YZ07_9VIRU Capsid protein OS=Common bean-associated gemycircularvirus OX=1897538 PE=4 SV=1
MM1 pKa = 7.75PVRR4 pKa = 11.84NRR6 pKa = 11.84NFPRR10 pKa = 11.84GWRR13 pKa = 11.84SRR15 pKa = 11.84YY16 pKa = 8.66RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 10.21NVLNITSRR27 pKa = 11.84KK28 pKa = 9.15KK29 pKa = 10.27QDD31 pKa = 3.19TMLPWTNATASSVSGGSTFTNQAAVFVGGNTYY63 pKa = 10.4ILPWCCTARR72 pKa = 11.84DD73 pKa = 3.85NILANGSPATLFSQSSRR90 pKa = 11.84TATQCYY96 pKa = 7.86MRR98 pKa = 11.84GLKK101 pKa = 9.88EE102 pKa = 4.22RR103 pKa = 11.84IQVQTSTGMPWQWRR117 pKa = 11.84RR118 pKa = 11.84ICFTLKK124 pKa = 10.67GDD126 pKa = 3.74TLTQYY131 pKa = 11.42NEE133 pKa = 3.63TAYY136 pKa = 9.89KK137 pKa = 10.12IIAEE141 pKa = 4.33NSSGWARR148 pKa = 11.84VVSNVGDD155 pKa = 3.69KK156 pKa = 11.45SNLAVAINNLVFRR169 pKa = 11.84GQVEE173 pKa = 4.5SDD175 pKa = 2.52WKK177 pKa = 11.13AFFNAKK183 pKa = 9.79VDD185 pKa = 3.87DD186 pKa = 4.09SRR188 pKa = 11.84VSLKK192 pKa = 10.16YY193 pKa = 10.65DD194 pKa = 2.81KK195 pKa = 10.44TIYY198 pKa = 10.49VRR200 pKa = 11.84TGNNFGSMRR209 pKa = 11.84DD210 pKa = 3.27YY211 pKa = 11.34HH212 pKa = 6.81FWHH215 pKa = 6.81KK216 pKa = 10.46MNSTLIYY223 pKa = 10.44DD224 pKa = 4.02DD225 pKa = 5.62DD226 pKa = 4.13EE227 pKa = 7.16AGVSMVPSYY236 pKa = 11.53YY237 pKa = 9.63STEE240 pKa = 3.96SRR242 pKa = 11.84QGMGDD247 pKa = 3.72YY248 pKa = 10.71YY249 pKa = 11.39VIDD252 pKa = 4.91IISAGTGATSSDD264 pKa = 2.86QMTFNPQATLYY275 pKa = 8.14WHH277 pKa = 6.53EE278 pKa = 4.17RR279 pKa = 3.47
MM1 pKa = 7.75PVRR4 pKa = 11.84NRR6 pKa = 11.84NFPRR10 pKa = 11.84GWRR13 pKa = 11.84SRR15 pKa = 11.84YY16 pKa = 8.66RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 10.21NVLNITSRR27 pKa = 11.84KK28 pKa = 9.15KK29 pKa = 10.27QDD31 pKa = 3.19TMLPWTNATASSVSGGSTFTNQAAVFVGGNTYY63 pKa = 10.4ILPWCCTARR72 pKa = 11.84DD73 pKa = 3.85NILANGSPATLFSQSSRR90 pKa = 11.84TATQCYY96 pKa = 7.86MRR98 pKa = 11.84GLKK101 pKa = 9.88EE102 pKa = 4.22RR103 pKa = 11.84IQVQTSTGMPWQWRR117 pKa = 11.84RR118 pKa = 11.84ICFTLKK124 pKa = 10.67GDD126 pKa = 3.74TLTQYY131 pKa = 11.42NEE133 pKa = 3.63TAYY136 pKa = 9.89KK137 pKa = 10.12IIAEE141 pKa = 4.33NSSGWARR148 pKa = 11.84VVSNVGDD155 pKa = 3.69KK156 pKa = 11.45SNLAVAINNLVFRR169 pKa = 11.84GQVEE173 pKa = 4.5SDD175 pKa = 2.52WKK177 pKa = 11.13AFFNAKK183 pKa = 9.79VDD185 pKa = 3.87DD186 pKa = 4.09SRR188 pKa = 11.84VSLKK192 pKa = 10.16YY193 pKa = 10.65DD194 pKa = 2.81KK195 pKa = 10.44TIYY198 pKa = 10.49VRR200 pKa = 11.84TGNNFGSMRR209 pKa = 11.84DD210 pKa = 3.27YY211 pKa = 11.34HH212 pKa = 6.81FWHH215 pKa = 6.81KK216 pKa = 10.46MNSTLIYY223 pKa = 10.44DD224 pKa = 4.02DD225 pKa = 5.62DD226 pKa = 4.13EE227 pKa = 7.16AGVSMVPSYY236 pKa = 11.53YY237 pKa = 9.63STEE240 pKa = 3.96SRR242 pKa = 11.84QGMGDD247 pKa = 3.72YY248 pKa = 10.71YY249 pKa = 11.39VIDD252 pKa = 4.91IISAGTGATSSDD264 pKa = 2.86QMTFNPQATLYY275 pKa = 8.14WHH277 pKa = 6.53EE278 pKa = 4.17RR279 pKa = 3.47
Molecular weight: 31.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
616 |
279 |
337 |
308.0 |
35.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.656 ± 0.095 | 1.786 ± 0.217 |
7.143 ± 1.09 | 5.032 ± 1.558 |
4.87 ± 0.572 | 8.117 ± 0.807 |
1.623 ± 0.338 | 4.383 ± 0.171 |
5.195 ± 0.552 | 6.006 ± 0.831 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.435 ± 0.488 | 5.519 ± 1.018 |
3.896 ± 0.635 | 2.922 ± 0.851 |
7.792 ± 0.164 | 6.494 ± 1.965 |
6.494 ± 1.523 | 5.519 ± 0.575 |
3.084 ± 0.087 | 5.032 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
