
Torque teno Leptonychotes weddellii virus-2
Taxonomy: Viruses; Anelloviridae; Lambdatorquevirus; Torque teno pinniped virus 9
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
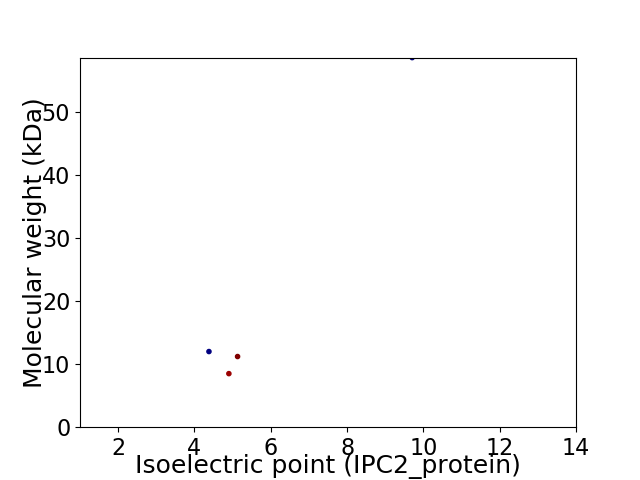
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
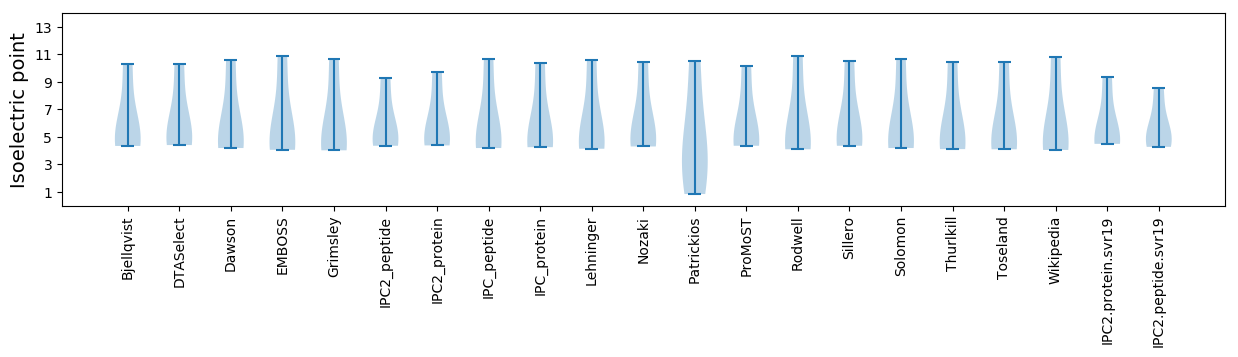
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2RW16|A0A1Z2RW16_9VIRU ORF3 OS=Torque teno Leptonychotes weddellii virus-2 OX=2012677 PE=4 SV=1
MM1 pKa = 7.52ALSNKK6 pKa = 8.91YY7 pKa = 10.0RR8 pKa = 11.84LAPYY12 pKa = 10.42SKK14 pKa = 10.86GPLRR18 pKa = 11.84LPTSFRR24 pKa = 11.84EE25 pKa = 4.17TSTQTDD31 pKa = 3.32ASKK34 pKa = 10.92KK35 pKa = 10.38ALFEE39 pKa = 4.01EE40 pKa = 4.47LLALIEE46 pKa = 4.89QDD48 pKa = 5.69SQPCWCQRR56 pKa = 11.84EE57 pKa = 4.06SDD59 pKa = 4.31LDD61 pKa = 4.01LEE63 pKa = 4.71SEE65 pKa = 4.33TDD67 pKa = 3.42SEE69 pKa = 4.79RR70 pKa = 11.84SIEE73 pKa = 3.97FLGSSWEE80 pKa = 4.14DD81 pKa = 3.38EE82 pKa = 4.32EE83 pKa = 5.44NPGGEE88 pKa = 4.57GPPLVPPEE96 pKa = 5.46GGAQWPSSFRR106 pKa = 11.84FF107 pKa = 3.43
MM1 pKa = 7.52ALSNKK6 pKa = 8.91YY7 pKa = 10.0RR8 pKa = 11.84LAPYY12 pKa = 10.42SKK14 pKa = 10.86GPLRR18 pKa = 11.84LPTSFRR24 pKa = 11.84EE25 pKa = 4.17TSTQTDD31 pKa = 3.32ASKK34 pKa = 10.92KK35 pKa = 10.38ALFEE39 pKa = 4.01EE40 pKa = 4.47LLALIEE46 pKa = 4.89QDD48 pKa = 5.69SQPCWCQRR56 pKa = 11.84EE57 pKa = 4.06SDD59 pKa = 4.31LDD61 pKa = 4.01LEE63 pKa = 4.71SEE65 pKa = 4.33TDD67 pKa = 3.42SEE69 pKa = 4.79RR70 pKa = 11.84SIEE73 pKa = 3.97FLGSSWEE80 pKa = 4.14DD81 pKa = 3.38EE82 pKa = 4.32EE83 pKa = 5.44NPGGEE88 pKa = 4.57GPPLVPPEE96 pKa = 5.46GGAQWPSSFRR106 pKa = 11.84FF107 pKa = 3.43
Molecular weight: 12.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RVZ1|A0A1Z2RVZ1_9VIRU ORF2 OS=Torque teno Leptonychotes weddellii virus-2 OX=2012677 PE=4 SV=1
MM1 pKa = 7.33ARR3 pKa = 11.84FWRR6 pKa = 11.84RR7 pKa = 11.84WRR9 pKa = 11.84KK10 pKa = 9.17RR11 pKa = 11.84PRR13 pKa = 11.84IWRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.83RR20 pKa = 11.84SYY22 pKa = 11.13RR23 pKa = 11.84PFRR26 pKa = 11.84RR27 pKa = 11.84WSHH30 pKa = 5.41RR31 pKa = 11.84RR32 pKa = 11.84WGRR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 5.82RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 5.44FRR42 pKa = 11.84HH43 pKa = 6.44RR44 pKa = 11.84YY45 pKa = 7.57AAVRR49 pKa = 11.84YY50 pKa = 8.91CPSRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 5.63KK57 pKa = 10.76KK58 pKa = 10.09LVVSGWEE65 pKa = 3.72PLGNVCATTTVCSEE79 pKa = 3.93AKK81 pKa = 9.45PYY83 pKa = 10.49KK84 pKa = 10.04ILEE87 pKa = 4.55CKK89 pKa = 9.99EE90 pKa = 4.54SEE92 pKa = 4.12IQQVANQMNKK102 pKa = 9.88DD103 pKa = 3.9GEE105 pKa = 4.61WNGSWGHH112 pKa = 5.88HH113 pKa = 4.66WFSFAGLLARR123 pKa = 11.84SQYY126 pKa = 10.45FFNTFSTDD134 pKa = 2.52WRR136 pKa = 11.84SYY138 pKa = 11.45DD139 pKa = 3.47YY140 pKa = 11.4LQFLGGYY147 pKa = 8.17VWLPRR152 pKa = 11.84QMYY155 pKa = 9.2LDD157 pKa = 2.99WMFYY161 pKa = 10.2IDD163 pKa = 5.95YY164 pKa = 10.6DD165 pKa = 4.03LQDD168 pKa = 3.45KK169 pKa = 8.54VTGEE173 pKa = 3.96KK174 pKa = 9.37MDD176 pKa = 3.38MYY178 pKa = 10.71KK179 pKa = 10.55NSKK182 pKa = 7.07TWFHH186 pKa = 6.57PAILLNRR193 pKa = 11.84RR194 pKa = 11.84GGIIVGSPHH203 pKa = 6.95RR204 pKa = 11.84FGIKK208 pKa = 9.3TMFRR212 pKa = 11.84RR213 pKa = 11.84LKK215 pKa = 10.45VKK217 pKa = 10.43PPANWEE223 pKa = 4.01GLYY226 pKa = 10.44NLPDD230 pKa = 3.96ALHH233 pKa = 6.68YY234 pKa = 10.85IFFHH238 pKa = 6.37WVWTWIDD245 pKa = 3.77LEE247 pKa = 4.21QSFMDD252 pKa = 3.99SDD254 pKa = 3.87WVKK257 pKa = 10.56HH258 pKa = 5.48IRR260 pKa = 11.84SGDD263 pKa = 3.18IGSQKK268 pKa = 9.97PCEE271 pKa = 4.1DD272 pKa = 3.75VPWFLGGIEE281 pKa = 4.35TKK283 pKa = 10.57QPTVQQGFDD292 pKa = 3.68KK293 pKa = 10.62RR294 pKa = 11.84ASWVDD299 pKa = 3.32RR300 pKa = 11.84EE301 pKa = 4.47KK302 pKa = 11.33YY303 pKa = 9.63CCKK306 pKa = 9.94KK307 pKa = 10.23QRR309 pKa = 11.84NFSDD313 pKa = 3.33PMDD316 pKa = 3.88NYY318 pKa = 10.61GRR320 pKa = 11.84WGPFCPYY327 pKa = 10.3VYY329 pKa = 10.84GMPYY333 pKa = 9.18NQSRR337 pKa = 11.84SLWFKK342 pKa = 10.47YY343 pKa = 10.42KK344 pKa = 10.69FFFKK348 pKa = 10.83LSGDD352 pKa = 3.6SVYY355 pKa = 10.58RR356 pKa = 11.84RR357 pKa = 11.84PPTSEE362 pKa = 4.15AEE364 pKa = 4.23EE365 pKa = 4.37LVPEE369 pKa = 5.41APGQQQNGPKK379 pKa = 9.39QQIPSSSILKK389 pKa = 10.36RR390 pKa = 11.84PLTPSDD396 pKa = 4.04ILPGDD401 pKa = 4.43LDD403 pKa = 3.83SDD405 pKa = 4.26GCIKK409 pKa = 10.38EE410 pKa = 4.21SPLRR414 pKa = 11.84RR415 pKa = 11.84IISSHH420 pKa = 5.37RR421 pKa = 11.84TGQPTLLVPKK431 pKa = 10.12RR432 pKa = 11.84VRR434 pKa = 11.84FRR436 pKa = 11.84LGKK439 pKa = 9.56RR440 pKa = 11.84DD441 pKa = 3.61RR442 pKa = 11.84QRR444 pKa = 11.84KK445 pKa = 5.01IHH447 pKa = 6.85RR448 pKa = 11.84ILRR451 pKa = 11.84QLMGGRR457 pKa = 11.84RR458 pKa = 11.84EE459 pKa = 4.4SGGGRR464 pKa = 11.84PPPGPPRR471 pKa = 11.84RR472 pKa = 11.84RR473 pKa = 11.84GPMALQLPLLAHH485 pKa = 6.31LHH487 pKa = 6.26ILPQQ491 pKa = 3.68
MM1 pKa = 7.33ARR3 pKa = 11.84FWRR6 pKa = 11.84RR7 pKa = 11.84WRR9 pKa = 11.84KK10 pKa = 9.17RR11 pKa = 11.84PRR13 pKa = 11.84IWRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.83RR20 pKa = 11.84SYY22 pKa = 11.13RR23 pKa = 11.84PFRR26 pKa = 11.84RR27 pKa = 11.84WSHH30 pKa = 5.41RR31 pKa = 11.84RR32 pKa = 11.84WGRR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 5.82RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 5.44FRR42 pKa = 11.84HH43 pKa = 6.44RR44 pKa = 11.84YY45 pKa = 7.57AAVRR49 pKa = 11.84YY50 pKa = 8.91CPSRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 5.63KK57 pKa = 10.76KK58 pKa = 10.09LVVSGWEE65 pKa = 3.72PLGNVCATTTVCSEE79 pKa = 3.93AKK81 pKa = 9.45PYY83 pKa = 10.49KK84 pKa = 10.04ILEE87 pKa = 4.55CKK89 pKa = 9.99EE90 pKa = 4.54SEE92 pKa = 4.12IQQVANQMNKK102 pKa = 9.88DD103 pKa = 3.9GEE105 pKa = 4.61WNGSWGHH112 pKa = 5.88HH113 pKa = 4.66WFSFAGLLARR123 pKa = 11.84SQYY126 pKa = 10.45FFNTFSTDD134 pKa = 2.52WRR136 pKa = 11.84SYY138 pKa = 11.45DD139 pKa = 3.47YY140 pKa = 11.4LQFLGGYY147 pKa = 8.17VWLPRR152 pKa = 11.84QMYY155 pKa = 9.2LDD157 pKa = 2.99WMFYY161 pKa = 10.2IDD163 pKa = 5.95YY164 pKa = 10.6DD165 pKa = 4.03LQDD168 pKa = 3.45KK169 pKa = 8.54VTGEE173 pKa = 3.96KK174 pKa = 9.37MDD176 pKa = 3.38MYY178 pKa = 10.71KK179 pKa = 10.55NSKK182 pKa = 7.07TWFHH186 pKa = 6.57PAILLNRR193 pKa = 11.84RR194 pKa = 11.84GGIIVGSPHH203 pKa = 6.95RR204 pKa = 11.84FGIKK208 pKa = 9.3TMFRR212 pKa = 11.84RR213 pKa = 11.84LKK215 pKa = 10.45VKK217 pKa = 10.43PPANWEE223 pKa = 4.01GLYY226 pKa = 10.44NLPDD230 pKa = 3.96ALHH233 pKa = 6.68YY234 pKa = 10.85IFFHH238 pKa = 6.37WVWTWIDD245 pKa = 3.77LEE247 pKa = 4.21QSFMDD252 pKa = 3.99SDD254 pKa = 3.87WVKK257 pKa = 10.56HH258 pKa = 5.48IRR260 pKa = 11.84SGDD263 pKa = 3.18IGSQKK268 pKa = 9.97PCEE271 pKa = 4.1DD272 pKa = 3.75VPWFLGGIEE281 pKa = 4.35TKK283 pKa = 10.57QPTVQQGFDD292 pKa = 3.68KK293 pKa = 10.62RR294 pKa = 11.84ASWVDD299 pKa = 3.32RR300 pKa = 11.84EE301 pKa = 4.47KK302 pKa = 11.33YY303 pKa = 9.63CCKK306 pKa = 9.94KK307 pKa = 10.23QRR309 pKa = 11.84NFSDD313 pKa = 3.33PMDD316 pKa = 3.88NYY318 pKa = 10.61GRR320 pKa = 11.84WGPFCPYY327 pKa = 10.3VYY329 pKa = 10.84GMPYY333 pKa = 9.18NQSRR337 pKa = 11.84SLWFKK342 pKa = 10.47YY343 pKa = 10.42KK344 pKa = 10.69FFFKK348 pKa = 10.83LSGDD352 pKa = 3.6SVYY355 pKa = 10.58RR356 pKa = 11.84RR357 pKa = 11.84PPTSEE362 pKa = 4.15AEE364 pKa = 4.23EE365 pKa = 4.37LVPEE369 pKa = 5.41APGQQQNGPKK379 pKa = 9.39QQIPSSSILKK389 pKa = 10.36RR390 pKa = 11.84PLTPSDD396 pKa = 4.04ILPGDD401 pKa = 4.43LDD403 pKa = 3.83SDD405 pKa = 4.26GCIKK409 pKa = 10.38EE410 pKa = 4.21SPLRR414 pKa = 11.84RR415 pKa = 11.84IISSHH420 pKa = 5.37RR421 pKa = 11.84TGQPTLLVPKK431 pKa = 10.12RR432 pKa = 11.84VRR434 pKa = 11.84FRR436 pKa = 11.84LGKK439 pKa = 9.56RR440 pKa = 11.84DD441 pKa = 3.61RR442 pKa = 11.84QRR444 pKa = 11.84KK445 pKa = 5.01IHH447 pKa = 6.85RR448 pKa = 11.84ILRR451 pKa = 11.84QLMGGRR457 pKa = 11.84RR458 pKa = 11.84EE459 pKa = 4.4SGGGRR464 pKa = 11.84PPPGPPRR471 pKa = 11.84RR472 pKa = 11.84RR473 pKa = 11.84GPMALQLPLLAHH485 pKa = 6.31LHH487 pKa = 6.26ILPQQ491 pKa = 3.68
Molecular weight: 58.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
779 |
73 |
491 |
194.8 |
22.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.979 ± 0.782 | 2.182 ± 0.649 |
5.52 ± 0.678 | 5.392 ± 1.935 |
5.006 ± 0.407 | 8.986 ± 2.194 |
2.696 ± 0.733 | 4.236 ± 0.531 |
5.648 ± 0.5 | 8.858 ± 2.767 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.439 ± 0.398 | 2.182 ± 0.441 |
6.932 ± 1.444 | 4.236 ± 0.912 |
8.858 ± 2.829 | 7.574 ± 1.655 |
3.979 ± 0.92 | 3.723 ± 0.798 |
3.723 ± 0.702 | 3.851 ± 0.628 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
