
Drosophila melanogaster (Fruit fly)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha;
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
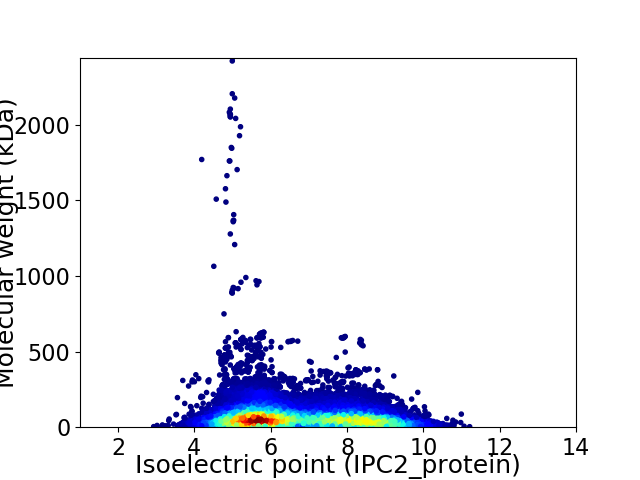
Virtual 2D-PAGE plot for 23524 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M3Q7U5|A0A6M3Q7U5_DROME Fish-lips isoform E OS=Drosophila melanogaster OX=7227 GN=Fili PE=4 SV=1
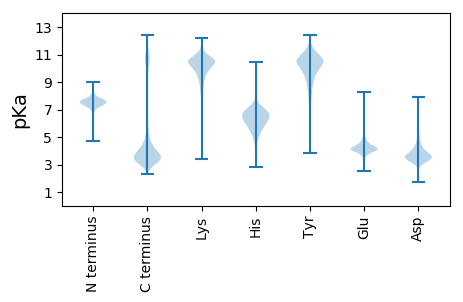
MM1 pKa = 7.6ILNTTSFLLILVCQAWGQSIKK22 pKa = 10.4IRR24 pKa = 11.84IPVWDD29 pKa = 4.06ASTIFSRR36 pKa = 11.84LTGIGINSGNTNNQNPSNQANNNQQNVEE64 pKa = 4.13YY65 pKa = 10.84NDD67 pKa = 3.64YY68 pKa = 11.38LNNYY72 pKa = 8.42YY73 pKa = 10.93NNYY76 pKa = 7.64WASAYY81 pKa = 10.58QNVANYY87 pKa = 10.31DD88 pKa = 3.44PYY90 pKa = 11.44GYY92 pKa = 9.95YY93 pKa = 10.13SPYY96 pKa = 10.39GPYY99 pKa = 10.5GSYY102 pKa = 11.28APGQYY107 pKa = 7.79QTITTPTVTQTTTPMVPTTTAVISTTEE134 pKa = 3.96TPICAVFPHH143 pKa = 6.89LPACQTSTSATSDD156 pKa = 3.35STTSSTPMTTSTTTTEE172 pKa = 4.64RR173 pKa = 11.84ITASTSSTTTTSTEE187 pKa = 3.76LSTTSTAIPTSTTSEE202 pKa = 4.27STSTAIPTSSTTDD215 pKa = 2.91STTTSTTSPSTEE227 pKa = 3.85STTTSTAIPTSTTSEE242 pKa = 4.27STSTAIPTSSTTDD255 pKa = 2.91STTTSTTSSSTEE267 pKa = 3.77STTTSTAIPTSTTSEE282 pKa = 4.27STSTAIPTSSTTDD295 pKa = 2.91STTTSTTSSSTEE307 pKa = 3.77STTTSTAIPTSTTSEE322 pKa = 4.27STSTAIPTSSTTDD335 pKa = 2.91STTTSTTSSSTEE347 pKa = 3.77STTTSTAIPTSTTSEE362 pKa = 4.27STSTAIPTSSTTDD375 pKa = 2.91STTTSTTSSSTEE387 pKa = 3.77STTTSTAIPTSTTSEE402 pKa = 4.28STSTALPTSSSTDD415 pKa = 2.99STTTSTTSSSTEE427 pKa = 3.77STTTSTAIPNSTTSEE442 pKa = 4.25STSTAIPTSSTTDD455 pKa = 2.91STTTSTTSSSTEE467 pKa = 3.77STTTSTAIPTSTTSEE482 pKa = 4.28STSTALPTSSSTDD495 pKa = 2.99STTTSTTSSSTEE507 pKa = 3.77STTTSTAIPNSTTSEE522 pKa = 4.25STSTAIPTSSTTDD535 pKa = 2.91STTTSTTSSSTEE547 pKa = 3.77STTTSTAIPTSTTSEE562 pKa = 4.27STSTAIPTSSTTDD575 pKa = 2.91STTTSTTSSPTEE587 pKa = 3.85STTTSTAIPTSTTSEE602 pKa = 4.27STSTAIPTSSTTDD615 pKa = 3.11STKK618 pKa = 10.66TSTTSSSTEE627 pKa = 3.7SSTTSTAIPTSTTSEE642 pKa = 4.28STSTALPTSSSTDD655 pKa = 2.99STTTSTTSSSTEE667 pKa = 3.77STTTSTAIPNSTTSEE682 pKa = 4.25STSTAIPTSSTTDD695 pKa = 2.91STTTSTTSSSTEE707 pKa = 3.77STTTSTAIPTSTTSEE722 pKa = 4.27STSTAIPTSSTTDD735 pKa = 2.91STTTSTTSSPTEE747 pKa = 3.85STTTSTAIPTSTTSEE762 pKa = 4.27STSTAIPTSSTTDD775 pKa = 2.91STTTSTTSSSTEE787 pKa = 3.7SSTTSTAIPTSSTSDD802 pKa = 3.26SSDD805 pKa = 3.06STTTSTASPSDD816 pKa = 3.37STTTSTEE823 pKa = 3.95SSTSTTTEE831 pKa = 3.8STSTSTAIPTSSTSDD846 pKa = 3.13STTTSTASPSDD857 pKa = 3.37STTTSTEE864 pKa = 3.95SSTSTTTEE872 pKa = 3.8STSTSTAIPTSSTSDD887 pKa = 3.13STTTSTASSSDD898 pKa = 3.28SATTSTAIPTSTTTGGISTSTAIPQTKK925 pKa = 8.55STDD928 pKa = 4.04SICNEE933 pKa = 3.88YY934 pKa = 10.56PMLPNCQSEE943 pKa = 4.42KK944 pKa = 10.84AVNLKK949 pKa = 10.45KK950 pKa = 10.17IEE952 pKa = 4.46SNMFEE957 pKa = 4.47TNPEE961 pKa = 3.76VSVFKK966 pKa = 10.7PEE968 pKa = 3.79QKK970 pKa = 9.64LTITSMPQSVTSLCHH985 pKa = 6.32FYY987 pKa = 10.59PRR989 pKa = 11.84LCLKK993 pKa = 10.49PEE995 pKa = 3.74EE996 pKa = 4.2AQFVRR1001 pKa = 11.84LADD1004 pKa = 4.08EE1005 pKa = 4.15YY1006 pKa = 11.48GKK1008 pKa = 9.67PLLLISSIEE1017 pKa = 3.9GRR1019 pKa = 11.84SVYY1022 pKa = 9.92AQKK1025 pKa = 11.04LPALWRR1031 pKa = 11.84AIRR1034 pKa = 11.84RR1035 pKa = 11.84THH1037 pKa = 6.44RR1038 pKa = 11.84NKK1040 pKa = 10.79
MM1 pKa = 7.6ILNTTSFLLILVCQAWGQSIKK22 pKa = 10.4IRR24 pKa = 11.84IPVWDD29 pKa = 4.06ASTIFSRR36 pKa = 11.84LTGIGINSGNTNNQNPSNQANNNQQNVEE64 pKa = 4.13YY65 pKa = 10.84NDD67 pKa = 3.64YY68 pKa = 11.38LNNYY72 pKa = 8.42YY73 pKa = 10.93NNYY76 pKa = 7.64WASAYY81 pKa = 10.58QNVANYY87 pKa = 10.31DD88 pKa = 3.44PYY90 pKa = 11.44GYY92 pKa = 9.95YY93 pKa = 10.13SPYY96 pKa = 10.39GPYY99 pKa = 10.5GSYY102 pKa = 11.28APGQYY107 pKa = 7.79QTITTPTVTQTTTPMVPTTTAVISTTEE134 pKa = 3.96TPICAVFPHH143 pKa = 6.89LPACQTSTSATSDD156 pKa = 3.35STTSSTPMTTSTTTTEE172 pKa = 4.64RR173 pKa = 11.84ITASTSSTTTTSTEE187 pKa = 3.76LSTTSTAIPTSTTSEE202 pKa = 4.27STSTAIPTSSTTDD215 pKa = 2.91STTTSTTSPSTEE227 pKa = 3.85STTTSTAIPTSTTSEE242 pKa = 4.27STSTAIPTSSTTDD255 pKa = 2.91STTTSTTSSSTEE267 pKa = 3.77STTTSTAIPTSTTSEE282 pKa = 4.27STSTAIPTSSTTDD295 pKa = 2.91STTTSTTSSSTEE307 pKa = 3.77STTTSTAIPTSTTSEE322 pKa = 4.27STSTAIPTSSTTDD335 pKa = 2.91STTTSTTSSSTEE347 pKa = 3.77STTTSTAIPTSTTSEE362 pKa = 4.27STSTAIPTSSTTDD375 pKa = 2.91STTTSTTSSSTEE387 pKa = 3.77STTTSTAIPTSTTSEE402 pKa = 4.28STSTALPTSSSTDD415 pKa = 2.99STTTSTTSSSTEE427 pKa = 3.77STTTSTAIPNSTTSEE442 pKa = 4.25STSTAIPTSSTTDD455 pKa = 2.91STTTSTTSSSTEE467 pKa = 3.77STTTSTAIPTSTTSEE482 pKa = 4.28STSTALPTSSSTDD495 pKa = 2.99STTTSTTSSSTEE507 pKa = 3.77STTTSTAIPNSTTSEE522 pKa = 4.25STSTAIPTSSTTDD535 pKa = 2.91STTTSTTSSSTEE547 pKa = 3.77STTTSTAIPTSTTSEE562 pKa = 4.27STSTAIPTSSTTDD575 pKa = 2.91STTTSTTSSPTEE587 pKa = 3.85STTTSTAIPTSTTSEE602 pKa = 4.27STSTAIPTSSTTDD615 pKa = 3.11STKK618 pKa = 10.66TSTTSSSTEE627 pKa = 3.7SSTTSTAIPTSTTSEE642 pKa = 4.28STSTALPTSSSTDD655 pKa = 2.99STTTSTTSSSTEE667 pKa = 3.77STTTSTAIPNSTTSEE682 pKa = 4.25STSTAIPTSSTTDD695 pKa = 2.91STTTSTTSSSTEE707 pKa = 3.77STTTSTAIPTSTTSEE722 pKa = 4.27STSTAIPTSSTTDD735 pKa = 2.91STTTSTTSSPTEE747 pKa = 3.85STTTSTAIPTSTTSEE762 pKa = 4.27STSTAIPTSSTTDD775 pKa = 2.91STTTSTTSSSTEE787 pKa = 3.7SSTTSTAIPTSSTSDD802 pKa = 3.26SSDD805 pKa = 3.06STTTSTASPSDD816 pKa = 3.37STTTSTEE823 pKa = 3.95SSTSTTTEE831 pKa = 3.8STSTSTAIPTSSTSDD846 pKa = 3.13STTTSTASPSDD857 pKa = 3.37STTTSTEE864 pKa = 3.95SSTSTTTEE872 pKa = 3.8STSTSTAIPTSSTSDD887 pKa = 3.13STTTSTASSSDD898 pKa = 3.28SATTSTAIPTSTTTGGISTSTAIPQTKK925 pKa = 8.55STDD928 pKa = 4.04SICNEE933 pKa = 3.88YY934 pKa = 10.56PMLPNCQSEE943 pKa = 4.42KK944 pKa = 10.84AVNLKK949 pKa = 10.45KK950 pKa = 10.17IEE952 pKa = 4.46SNMFEE957 pKa = 4.47TNPEE961 pKa = 3.76VSVFKK966 pKa = 10.7PEE968 pKa = 3.79QKK970 pKa = 9.64LTITSMPQSVTSLCHH985 pKa = 6.32FYY987 pKa = 10.59PRR989 pKa = 11.84LCLKK993 pKa = 10.49PEE995 pKa = 3.74EE996 pKa = 4.2AQFVRR1001 pKa = 11.84LADD1004 pKa = 4.08EE1005 pKa = 4.15YY1006 pKa = 11.48GKK1008 pKa = 9.67PLLLISSIEE1017 pKa = 3.9GRR1019 pKa = 11.84SVYY1022 pKa = 9.92AQKK1025 pKa = 11.04LPALWRR1031 pKa = 11.84AIRR1034 pKa = 11.84RR1035 pKa = 11.84THH1037 pKa = 6.44RR1038 pKa = 11.84NKK1040 pKa = 10.79
Molecular weight: 105.18 kDa
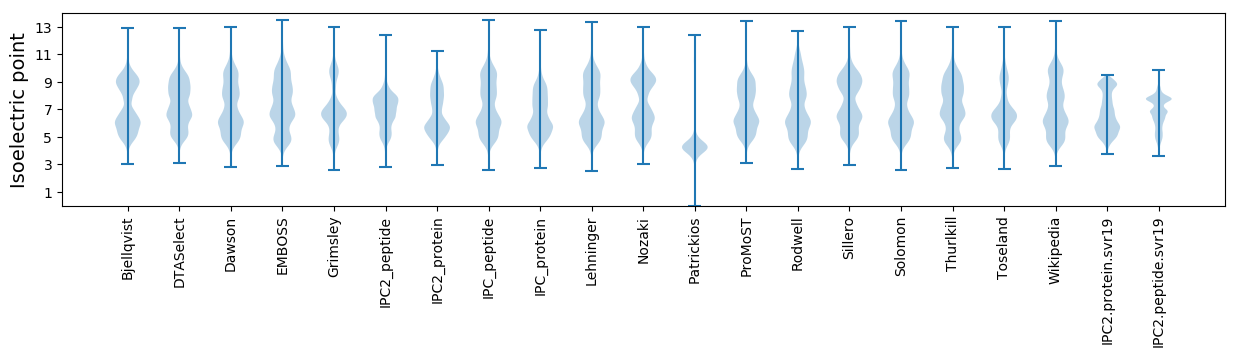
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9V399|CP303_DROME Probable cytochrome P450 303a1 OS=Drosophila melanogaster OX=7227 GN=Cyp303a1 PE=2 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 7.85RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.6MRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 11.11
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 7.85RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.6MRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 11.11
Molecular weight: 3.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
16289205 |
11 |
22949 |
692.5 |
76.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.421 ± 0.022 | 1.918 ± 0.049 |
5.196 ± 0.015 | 6.582 ± 0.039 |
3.293 ± 0.016 | 6.21 ± 0.022 |
2.644 ± 0.01 | 4.797 ± 0.015 |
5.504 ± 0.029 | 8.717 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.237 ± 0.014 | 4.735 ± 0.014 |
5.764 ± 0.038 | 5.455 ± 0.024 |
5.461 ± 0.017 | 8.61 ± 0.032 |
5.872 ± 0.027 | 5.864 ± 0.019 |
0.917 ± 0.007 | 2.799 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
