
Castellaniella defragrans 65Phen
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Alcaligenaceae; Castellaniella; Castellaniella defragrans
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
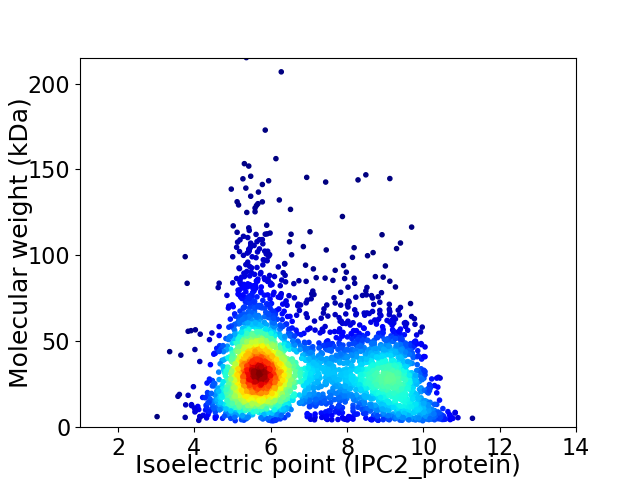
Virtual 2D-PAGE plot for 3575 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
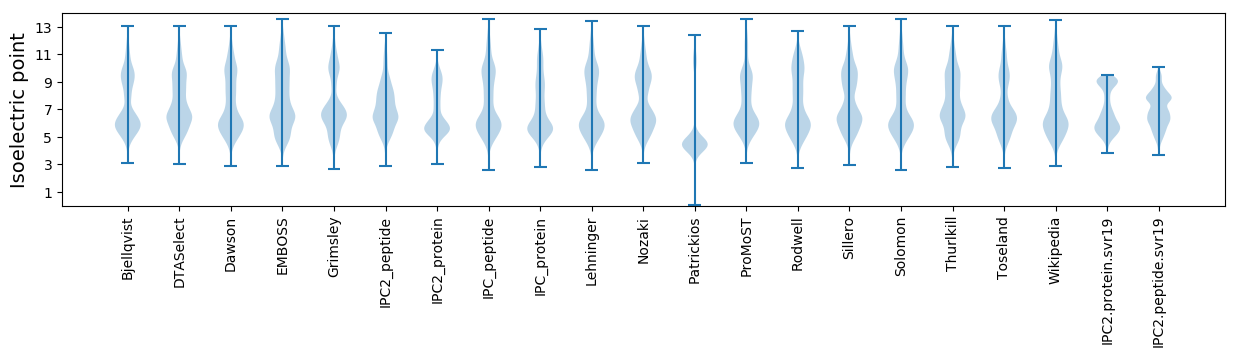
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8X940|W8X940_CASDE Protocatechuate 4 5-dioxygenase beta chain OS=Castellaniella defragrans 65Phen OX=1437824 GN=BN940_08486 PE=4 SV=1
MM1 pKa = 7.88ALPQWTDD8 pKa = 3.16QQVFDD13 pKa = 4.19QMNSGSTWTSSLITYY28 pKa = 8.16AFPQSSSVFDD38 pKa = 4.29PDD40 pKa = 3.99LAEE43 pKa = 4.41HH44 pKa = 6.56FAGFSPLNTAQQPLVHH60 pKa = 6.77LAMMLWDD67 pKa = 4.6DD68 pKa = 5.44LIARR72 pKa = 11.84DD73 pKa = 3.66IVQGSVHH80 pKa = 6.96DD81 pKa = 4.36ADD83 pKa = 4.2IMLSNTSSTDD93 pKa = 2.92GYY95 pKa = 11.33AFAVSGGTVWFSSKK109 pKa = 10.59EE110 pKa = 3.94DD111 pKa = 3.68LLQSPEE117 pKa = 3.64IGKK120 pKa = 10.17DD121 pKa = 3.37GFVTFLHH128 pKa = 6.83EE129 pKa = 4.39IGHH132 pKa = 6.39ALGLNHH138 pKa = 6.48MGDD141 pKa = 4.18YY142 pKa = 10.85NGEE145 pKa = 4.2DD146 pKa = 3.78DD147 pKa = 5.12NGPSSYY153 pKa = 10.46QDD155 pKa = 2.99SDD157 pKa = 3.5MLSIMSYY164 pKa = 10.32YY165 pKa = 10.03GPGMNGGKK173 pKa = 10.41GLVAWGDD180 pKa = 3.45WFASDD185 pKa = 4.4IGSEE189 pKa = 4.8GYY191 pKa = 10.2SPQTPMVNDD200 pKa = 3.14IMVIQRR206 pKa = 11.84LYY208 pKa = 11.16GADD211 pKa = 3.04TTTRR215 pKa = 11.84EE216 pKa = 4.07GNTVYY221 pKa = 10.85GFGSNIQGPLAQIYY235 pKa = 10.15DD236 pKa = 3.77FSINQHH242 pKa = 6.97PILTIYY248 pKa = 10.45DD249 pKa = 3.53AGGVDD254 pKa = 4.69TINLSGWGTDD264 pKa = 3.54SLIDD268 pKa = 4.41LNPGQYY274 pKa = 10.6SSVNGMTNNLAIAKK288 pKa = 7.64ATLIEE293 pKa = 4.12NAVAGAGNDD302 pKa = 3.56VLIGNSADD310 pKa = 3.19NHH312 pKa = 6.45LDD314 pKa = 3.24GGAGQDD320 pKa = 3.0SAMFSGALPGYY331 pKa = 9.59DD332 pKa = 3.23LSYY335 pKa = 11.27DD336 pKa = 3.71VFSRR340 pKa = 11.84EE341 pKa = 3.86YY342 pKa = 10.23TVVDD346 pKa = 3.52MTAGRR351 pKa = 11.84DD352 pKa = 3.82GTDD355 pKa = 2.87TLINIEE361 pKa = 4.03YY362 pKa = 10.77ANFNGAGGDD371 pKa = 4.07LNDD374 pKa = 4.55LTPAVYY380 pKa = 10.18RR381 pKa = 11.84FYY383 pKa = 11.29NAGLGLHH390 pKa = 7.23FYY392 pKa = 10.05TSNNDD397 pKa = 2.62EE398 pKa = 4.2ATAVTRR404 pKa = 11.84MNGFVYY410 pKa = 10.24EE411 pKa = 4.23GVGFGRR417 pKa = 11.84SVEE420 pKa = 4.25GAGIPDD426 pKa = 4.49ADD428 pKa = 3.74TVAVQRR434 pKa = 11.84FYY436 pKa = 11.68NQATGDD442 pKa = 3.54HH443 pKa = 7.26FYY445 pKa = 10.41TAGADD450 pKa = 3.8EE451 pKa = 4.87ARR453 pKa = 11.84HH454 pKa = 5.21LMEE457 pKa = 6.67IGGAWQYY464 pKa = 10.88EE465 pKa = 3.92GRR467 pKa = 11.84AFNAYY472 pKa = 7.09GTQADD477 pKa = 4.43GTTALYY483 pKa = 10.59RR484 pKa = 11.84FLNTEE489 pKa = 4.18SGTHH493 pKa = 7.12FYY495 pKa = 9.87TADD498 pKa = 3.3IAEE501 pKa = 4.83KK502 pKa = 9.53EE503 pKa = 4.2AVMQSGYY510 pKa = 10.66FSYY513 pKa = 10.74EE514 pKa = 3.59GIAFYY519 pKa = 9.76VTAA522 pKa = 5.13
MM1 pKa = 7.88ALPQWTDD8 pKa = 3.16QQVFDD13 pKa = 4.19QMNSGSTWTSSLITYY28 pKa = 8.16AFPQSSSVFDD38 pKa = 4.29PDD40 pKa = 3.99LAEE43 pKa = 4.41HH44 pKa = 6.56FAGFSPLNTAQQPLVHH60 pKa = 6.77LAMMLWDD67 pKa = 4.6DD68 pKa = 5.44LIARR72 pKa = 11.84DD73 pKa = 3.66IVQGSVHH80 pKa = 6.96DD81 pKa = 4.36ADD83 pKa = 4.2IMLSNTSSTDD93 pKa = 2.92GYY95 pKa = 11.33AFAVSGGTVWFSSKK109 pKa = 10.59EE110 pKa = 3.94DD111 pKa = 3.68LLQSPEE117 pKa = 3.64IGKK120 pKa = 10.17DD121 pKa = 3.37GFVTFLHH128 pKa = 6.83EE129 pKa = 4.39IGHH132 pKa = 6.39ALGLNHH138 pKa = 6.48MGDD141 pKa = 4.18YY142 pKa = 10.85NGEE145 pKa = 4.2DD146 pKa = 3.78DD147 pKa = 5.12NGPSSYY153 pKa = 10.46QDD155 pKa = 2.99SDD157 pKa = 3.5MLSIMSYY164 pKa = 10.32YY165 pKa = 10.03GPGMNGGKK173 pKa = 10.41GLVAWGDD180 pKa = 3.45WFASDD185 pKa = 4.4IGSEE189 pKa = 4.8GYY191 pKa = 10.2SPQTPMVNDD200 pKa = 3.14IMVIQRR206 pKa = 11.84LYY208 pKa = 11.16GADD211 pKa = 3.04TTTRR215 pKa = 11.84EE216 pKa = 4.07GNTVYY221 pKa = 10.85GFGSNIQGPLAQIYY235 pKa = 10.15DD236 pKa = 3.77FSINQHH242 pKa = 6.97PILTIYY248 pKa = 10.45DD249 pKa = 3.53AGGVDD254 pKa = 4.69TINLSGWGTDD264 pKa = 3.54SLIDD268 pKa = 4.41LNPGQYY274 pKa = 10.6SSVNGMTNNLAIAKK288 pKa = 7.64ATLIEE293 pKa = 4.12NAVAGAGNDD302 pKa = 3.56VLIGNSADD310 pKa = 3.19NHH312 pKa = 6.45LDD314 pKa = 3.24GGAGQDD320 pKa = 3.0SAMFSGALPGYY331 pKa = 9.59DD332 pKa = 3.23LSYY335 pKa = 11.27DD336 pKa = 3.71VFSRR340 pKa = 11.84EE341 pKa = 3.86YY342 pKa = 10.23TVVDD346 pKa = 3.52MTAGRR351 pKa = 11.84DD352 pKa = 3.82GTDD355 pKa = 2.87TLINIEE361 pKa = 4.03YY362 pKa = 10.77ANFNGAGGDD371 pKa = 4.07LNDD374 pKa = 4.55LTPAVYY380 pKa = 10.18RR381 pKa = 11.84FYY383 pKa = 11.29NAGLGLHH390 pKa = 7.23FYY392 pKa = 10.05TSNNDD397 pKa = 2.62EE398 pKa = 4.2ATAVTRR404 pKa = 11.84MNGFVYY410 pKa = 10.24EE411 pKa = 4.23GVGFGRR417 pKa = 11.84SVEE420 pKa = 4.25GAGIPDD426 pKa = 4.49ADD428 pKa = 3.74TVAVQRR434 pKa = 11.84FYY436 pKa = 11.68NQATGDD442 pKa = 3.54HH443 pKa = 7.26FYY445 pKa = 10.41TAGADD450 pKa = 3.8EE451 pKa = 4.87ARR453 pKa = 11.84HH454 pKa = 5.21LMEE457 pKa = 6.67IGGAWQYY464 pKa = 10.88EE465 pKa = 3.92GRR467 pKa = 11.84AFNAYY472 pKa = 7.09GTQADD477 pKa = 4.43GTTALYY483 pKa = 10.59RR484 pKa = 11.84FLNTEE489 pKa = 4.18SGTHH493 pKa = 7.12FYY495 pKa = 9.87TADD498 pKa = 3.3IAEE501 pKa = 4.83KK502 pKa = 9.53EE503 pKa = 4.2AVMQSGYY510 pKa = 10.66FSYY513 pKa = 10.74EE514 pKa = 3.59GIAFYY519 pKa = 9.76VTAA522 pKa = 5.13
Molecular weight: 56.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8X0C6|W8X0C6_CASDE DNA polymerase IV OS=Castellaniella defragrans 65Phen OX=1437824 GN=dinB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.99TRR25 pKa = 11.84AGRR28 pKa = 11.84AVLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.99TRR25 pKa = 11.84AGRR28 pKa = 11.84AVLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1162262 |
37 |
1991 |
325.1 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.341 ± 0.064 | 0.992 ± 0.015 |
5.632 ± 0.029 | 5.097 ± 0.037 |
3.327 ± 0.028 | 8.823 ± 0.042 |
2.317 ± 0.021 | 4.482 ± 0.031 |
2.33 ± 0.03 | 11.124 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.381 ± 0.019 | 2.168 ± 0.023 |
5.583 ± 0.036 | 3.756 ± 0.029 |
8.099 ± 0.047 | 4.918 ± 0.031 |
4.661 ± 0.028 | 7.172 ± 0.033 |
1.526 ± 0.018 | 2.272 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
