
Scleropages formosus (Asian bonytongue) (Osteoglossum formosum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Osteoglossocephala;
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
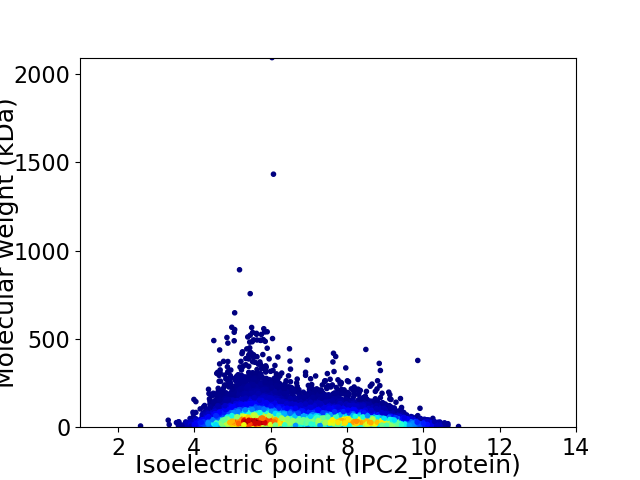
Virtual 2D-PAGE plot for 24200 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P7XQ50|A0A0P7XQ50_SCLFO Uncharacterized protein OS=Scleropages formosus OX=113540 GN=Z043_101413 PE=4 SV=1
II1 pKa = 7.96KK2 pKa = 10.27EE3 pKa = 4.1IIFLKK8 pKa = 8.52NTIMEE13 pKa = 4.42CEE15 pKa = 4.03ACGMGNDD22 pKa = 4.29KK23 pKa = 8.76PTHH26 pKa = 6.1PSCVPNPCHH35 pKa = 6.51PGVMCMEE42 pKa = 4.19TAGGIKK48 pKa = 10.08CGPCPEE54 pKa = 4.18GTEE57 pKa = 4.59GNGIHH62 pKa = 6.17CTDD65 pKa = 3.76VDD67 pKa = 3.73EE68 pKa = 5.84CLTKK72 pKa = 10.33PCHH75 pKa = 6.12TGVRR79 pKa = 11.84CINTFPGFRR88 pKa = 11.84CGPCPAGFTGPVVQGIGLSYY108 pKa = 11.24ARR110 pKa = 11.84TNKK113 pKa = 9.79QGSFHH118 pKa = 7.18CGPCKK123 pKa = 10.07PGFIGDD129 pKa = 3.6QQRR132 pKa = 11.84GCKK135 pKa = 9.66AEE137 pKa = 4.19KK138 pKa = 10.71ACGTDD143 pKa = 3.76QLNLCHH149 pKa = 6.56TNAEE153 pKa = 4.73CIVHH157 pKa = 6.84RR158 pKa = 11.84DD159 pKa = 3.33GSIEE163 pKa = 4.27CVCGVGWAGNGYY175 pKa = 9.93LCGLDD180 pKa = 3.18IDD182 pKa = 4.57IDD184 pKa = 3.96GFPDD188 pKa = 3.28EE189 pKa = 5.37KK190 pKa = 11.21LNCPEE195 pKa = 4.13TNCAKK200 pKa = 10.52DD201 pKa = 3.25NCLTVPNSGQEE212 pKa = 3.86DD213 pKa = 4.02ADD215 pKa = 4.12RR216 pKa = 11.84DD217 pKa = 4.5GIGDD221 pKa = 4.0ACDD224 pKa = 3.53EE225 pKa = 4.74DD226 pKa = 5.68ADD228 pKa = 4.2GDD230 pKa = 4.54GIINTEE236 pKa = 4.36DD237 pKa = 3.41NCVLKK242 pKa = 10.98PNVNQRR248 pKa = 11.84NVDD251 pKa = 3.21QDD253 pKa = 4.02DD254 pKa = 4.43FGDD257 pKa = 3.82ACDD260 pKa = 3.59NCRR263 pKa = 11.84LIINNDD269 pKa = 3.34QKK271 pKa = 11.27DD272 pKa = 3.3TDD274 pKa = 4.15ADD276 pKa = 4.15SKK278 pKa = 11.83GDD280 pKa = 3.68ACDD283 pKa = 4.97DD284 pKa = 5.26DD285 pKa = 5.19IDD287 pKa = 6.24GDD289 pKa = 4.41GIPNNVDD296 pKa = 2.97NCVMVPNRR304 pKa = 11.84DD305 pKa = 3.32QKK307 pKa = 11.54DD308 pKa = 3.36RR309 pKa = 11.84DD310 pKa = 3.49GDD312 pKa = 4.3GVGDD316 pKa = 4.14ACDD319 pKa = 3.15SCPYY323 pKa = 9.76VRR325 pKa = 11.84NPDD328 pKa = 3.61QLDD331 pKa = 3.29VDD333 pKa = 4.07NDD335 pKa = 4.78LIGDD339 pKa = 3.96PCDD342 pKa = 3.62TNKK345 pKa = 10.93DD346 pKa = 3.49SDD348 pKa = 4.16GDD350 pKa = 3.81GHH352 pKa = 6.91QDD354 pKa = 4.2SRR356 pKa = 11.84DD357 pKa = 3.47NCPAVINSSQLDD369 pKa = 3.54TDD371 pKa = 3.76RR372 pKa = 11.84DD373 pKa = 4.22GVGDD377 pKa = 4.09EE378 pKa = 5.64CDD380 pKa = 5.36DD381 pKa = 5.71DD382 pKa = 6.34DD383 pKa = 7.03DD384 pKa = 6.2NDD386 pKa = 5.25GIPDD390 pKa = 4.77LFPPGPDD397 pKa = 2.86NCRR400 pKa = 11.84LVPNPLQEE408 pKa = 5.49DD409 pKa = 3.7SDD411 pKa = 4.43GDD413 pKa = 4.19GIGNVCEE420 pKa = 4.02NDD422 pKa = 3.15YY423 pKa = 11.84DD424 pKa = 4.22NDD426 pKa = 4.18TFVDD430 pKa = 4.53MIDD433 pKa = 3.5VCPEE437 pKa = 3.68NAEE440 pKa = 4.04VTLTDD445 pKa = 3.96FRR447 pKa = 11.84AFQTVVLDD455 pKa = 4.1PEE457 pKa = 5.32GDD459 pKa = 3.65AQIDD463 pKa = 4.13PNWVVLNQGRR473 pKa = 11.84EE474 pKa = 3.9IVQTMNSDD482 pKa = 3.19PGLAVGYY489 pKa = 7.31TAFNGVDD496 pKa = 3.99FEE498 pKa = 4.97GTFHH502 pKa = 6.81VNTVTDD508 pKa = 3.57DD509 pKa = 4.08DD510 pKa = 4.35YY511 pKa = 12.0AGFIFGYY518 pKa = 9.58QDD520 pKa = 2.83SSSFYY525 pKa = 9.91VVMWKK530 pKa = 9.52QVEE533 pKa = 4.18QIYY536 pKa = 8.33WQANPFRR543 pKa = 11.84AVAEE547 pKa = 4.08PGIQLKK553 pKa = 10.06AVKK556 pKa = 10.35SNTGPGEE563 pKa = 3.91NLRR566 pKa = 11.84NSLWHH571 pKa = 6.45TGDD574 pKa = 3.39SNGQVRR580 pKa = 11.84LLWKK584 pKa = 10.35DD585 pKa = 3.22PRR587 pKa = 11.84NVGWKK592 pKa = 10.34DD593 pKa = 3.02KK594 pKa = 10.83TSYY597 pKa = 10.52RR598 pKa = 11.84WFLQHH603 pKa = 6.88RR604 pKa = 11.84PHH606 pKa = 7.4DD607 pKa = 4.02GYY609 pKa = 10.74IRR611 pKa = 11.84VRR613 pKa = 11.84FYY615 pKa = 11.04EE616 pKa = 4.86GPQMVADD623 pKa = 3.92SGIIIDD629 pKa = 3.57TTMRR633 pKa = 11.84GGRR636 pKa = 11.84LGVFCFSQEE645 pKa = 3.94NIIWSNLRR653 pKa = 11.84YY654 pKa = 9.68RR655 pKa = 11.84CNDD658 pKa = 3.2TTPEE662 pKa = 4.15DD663 pKa = 3.77FDD665 pKa = 3.94TYY667 pKa = 10.59RR668 pKa = 11.84IQQIQLQAA676 pKa = 3.49
II1 pKa = 7.96KK2 pKa = 10.27EE3 pKa = 4.1IIFLKK8 pKa = 8.52NTIMEE13 pKa = 4.42CEE15 pKa = 4.03ACGMGNDD22 pKa = 4.29KK23 pKa = 8.76PTHH26 pKa = 6.1PSCVPNPCHH35 pKa = 6.51PGVMCMEE42 pKa = 4.19TAGGIKK48 pKa = 10.08CGPCPEE54 pKa = 4.18GTEE57 pKa = 4.59GNGIHH62 pKa = 6.17CTDD65 pKa = 3.76VDD67 pKa = 3.73EE68 pKa = 5.84CLTKK72 pKa = 10.33PCHH75 pKa = 6.12TGVRR79 pKa = 11.84CINTFPGFRR88 pKa = 11.84CGPCPAGFTGPVVQGIGLSYY108 pKa = 11.24ARR110 pKa = 11.84TNKK113 pKa = 9.79QGSFHH118 pKa = 7.18CGPCKK123 pKa = 10.07PGFIGDD129 pKa = 3.6QQRR132 pKa = 11.84GCKK135 pKa = 9.66AEE137 pKa = 4.19KK138 pKa = 10.71ACGTDD143 pKa = 3.76QLNLCHH149 pKa = 6.56TNAEE153 pKa = 4.73CIVHH157 pKa = 6.84RR158 pKa = 11.84DD159 pKa = 3.33GSIEE163 pKa = 4.27CVCGVGWAGNGYY175 pKa = 9.93LCGLDD180 pKa = 3.18IDD182 pKa = 4.57IDD184 pKa = 3.96GFPDD188 pKa = 3.28EE189 pKa = 5.37KK190 pKa = 11.21LNCPEE195 pKa = 4.13TNCAKK200 pKa = 10.52DD201 pKa = 3.25NCLTVPNSGQEE212 pKa = 3.86DD213 pKa = 4.02ADD215 pKa = 4.12RR216 pKa = 11.84DD217 pKa = 4.5GIGDD221 pKa = 4.0ACDD224 pKa = 3.53EE225 pKa = 4.74DD226 pKa = 5.68ADD228 pKa = 4.2GDD230 pKa = 4.54GIINTEE236 pKa = 4.36DD237 pKa = 3.41NCVLKK242 pKa = 10.98PNVNQRR248 pKa = 11.84NVDD251 pKa = 3.21QDD253 pKa = 4.02DD254 pKa = 4.43FGDD257 pKa = 3.82ACDD260 pKa = 3.59NCRR263 pKa = 11.84LIINNDD269 pKa = 3.34QKK271 pKa = 11.27DD272 pKa = 3.3TDD274 pKa = 4.15ADD276 pKa = 4.15SKK278 pKa = 11.83GDD280 pKa = 3.68ACDD283 pKa = 4.97DD284 pKa = 5.26DD285 pKa = 5.19IDD287 pKa = 6.24GDD289 pKa = 4.41GIPNNVDD296 pKa = 2.97NCVMVPNRR304 pKa = 11.84DD305 pKa = 3.32QKK307 pKa = 11.54DD308 pKa = 3.36RR309 pKa = 11.84DD310 pKa = 3.49GDD312 pKa = 4.3GVGDD316 pKa = 4.14ACDD319 pKa = 3.15SCPYY323 pKa = 9.76VRR325 pKa = 11.84NPDD328 pKa = 3.61QLDD331 pKa = 3.29VDD333 pKa = 4.07NDD335 pKa = 4.78LIGDD339 pKa = 3.96PCDD342 pKa = 3.62TNKK345 pKa = 10.93DD346 pKa = 3.49SDD348 pKa = 4.16GDD350 pKa = 3.81GHH352 pKa = 6.91QDD354 pKa = 4.2SRR356 pKa = 11.84DD357 pKa = 3.47NCPAVINSSQLDD369 pKa = 3.54TDD371 pKa = 3.76RR372 pKa = 11.84DD373 pKa = 4.22GVGDD377 pKa = 4.09EE378 pKa = 5.64CDD380 pKa = 5.36DD381 pKa = 5.71DD382 pKa = 6.34DD383 pKa = 7.03DD384 pKa = 6.2NDD386 pKa = 5.25GIPDD390 pKa = 4.77LFPPGPDD397 pKa = 2.86NCRR400 pKa = 11.84LVPNPLQEE408 pKa = 5.49DD409 pKa = 3.7SDD411 pKa = 4.43GDD413 pKa = 4.19GIGNVCEE420 pKa = 4.02NDD422 pKa = 3.15YY423 pKa = 11.84DD424 pKa = 4.22NDD426 pKa = 4.18TFVDD430 pKa = 4.53MIDD433 pKa = 3.5VCPEE437 pKa = 3.68NAEE440 pKa = 4.04VTLTDD445 pKa = 3.96FRR447 pKa = 11.84AFQTVVLDD455 pKa = 4.1PEE457 pKa = 5.32GDD459 pKa = 3.65AQIDD463 pKa = 4.13PNWVVLNQGRR473 pKa = 11.84EE474 pKa = 3.9IVQTMNSDD482 pKa = 3.19PGLAVGYY489 pKa = 7.31TAFNGVDD496 pKa = 3.99FEE498 pKa = 4.97GTFHH502 pKa = 6.81VNTVTDD508 pKa = 3.57DD509 pKa = 4.08DD510 pKa = 4.35YY511 pKa = 12.0AGFIFGYY518 pKa = 9.58QDD520 pKa = 2.83SSSFYY525 pKa = 9.91VVMWKK530 pKa = 9.52QVEE533 pKa = 4.18QIYY536 pKa = 8.33WQANPFRR543 pKa = 11.84AVAEE547 pKa = 4.08PGIQLKK553 pKa = 10.06AVKK556 pKa = 10.35SNTGPGEE563 pKa = 3.91NLRR566 pKa = 11.84NSLWHH571 pKa = 6.45TGDD574 pKa = 3.39SNGQVRR580 pKa = 11.84LLWKK584 pKa = 10.35DD585 pKa = 3.22PRR587 pKa = 11.84NVGWKK592 pKa = 10.34DD593 pKa = 3.02KK594 pKa = 10.83TSYY597 pKa = 10.52RR598 pKa = 11.84WFLQHH603 pKa = 6.88RR604 pKa = 11.84PHH606 pKa = 7.4DD607 pKa = 4.02GYY609 pKa = 10.74IRR611 pKa = 11.84VRR613 pKa = 11.84FYY615 pKa = 11.04EE616 pKa = 4.86GPQMVADD623 pKa = 3.92SGIIIDD629 pKa = 3.57TTMRR633 pKa = 11.84GGRR636 pKa = 11.84LGVFCFSQEE645 pKa = 3.94NIIWSNLRR653 pKa = 11.84YY654 pKa = 9.68RR655 pKa = 11.84CNDD658 pKa = 3.2TTPEE662 pKa = 4.15DD663 pKa = 3.77FDD665 pKa = 3.94TYY667 pKa = 10.59RR668 pKa = 11.84IQQIQLQAA676 pKa = 3.49
Molecular weight: 73.96 kDa
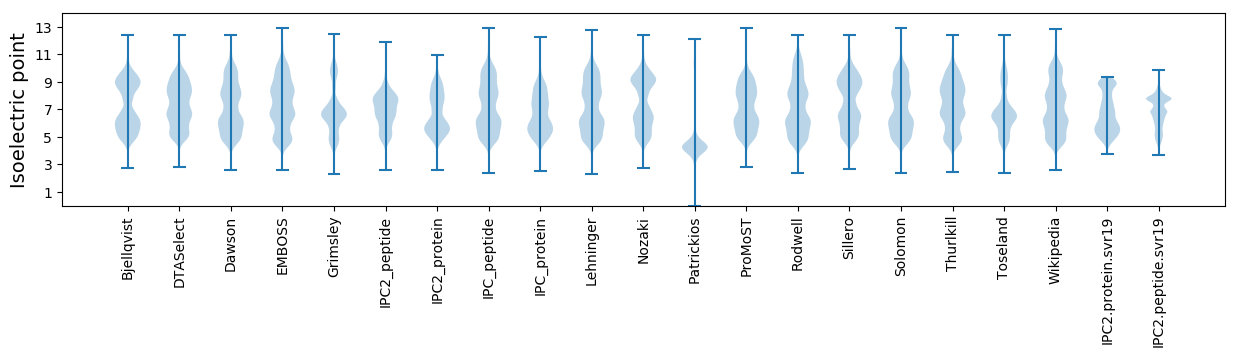
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P7Y0A6|A0A0P7Y0A6_SCLFO KN motif and ankyrin repeat domain-containing protein 2-like (Fragment) OS=Scleropages formosus OX=113540 GN=Z043_123050 PE=4 SV=1
MM1 pKa = 7.49PRR3 pKa = 11.84SFMWAPDD10 pKa = 3.59PQAWLRR16 pKa = 11.84DD17 pKa = 3.89DD18 pKa = 4.78AVPTALAVLWAAMALEE34 pKa = 4.71GRR36 pKa = 11.84CGEE39 pKa = 4.42RR40 pKa = 11.84GATPARR46 pKa = 11.84RR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84QAPVPRR58 pKa = 11.84TRR60 pKa = 11.84LHH62 pKa = 6.47GLRR65 pKa = 11.84HH66 pKa = 4.37VCARR70 pKa = 11.84GSSIGRR76 pKa = 11.84RR77 pKa = 11.84TFWILALCTSLGLLLSWSSNRR98 pKa = 11.84LLHH101 pKa = 6.06WLSFPTYY108 pKa = 8.99TKK110 pKa = 10.87VHH112 pKa = 5.47TEE114 pKa = 3.7WAKK117 pKa = 10.13EE118 pKa = 3.91LPFPTVTICNNNPVRR133 pKa = 11.84FYY135 pKa = 11.58KK136 pKa = 9.61LTKK139 pKa = 9.75SDD141 pKa = 4.41LYY143 pKa = 11.08FAGHH147 pKa = 6.37WLGLLLANRR156 pKa = 11.84TARR159 pKa = 11.84PLVLDD164 pKa = 4.46LLQEE168 pKa = 4.02DD169 pKa = 3.71RR170 pKa = 11.84RR171 pKa = 11.84AWFHH175 pKa = 6.32KK176 pKa = 10.68LSDD179 pKa = 3.79FRR181 pKa = 11.84LFLPPRR187 pKa = 11.84HH188 pKa = 6.13FEE190 pKa = 4.03GTNLEE195 pKa = 4.16FMDD198 pKa = 4.46RR199 pKa = 11.84LSHH202 pKa = 6.68QLDD205 pKa = 4.24DD206 pKa = 4.07MLLSCKK212 pKa = 10.06YY213 pKa = 9.52RR214 pKa = 11.84GEE216 pKa = 4.02MCGPQNFSTATRR228 pKa = 11.84GRR230 pKa = 11.84VAAGNKK236 pKa = 9.94DD237 pKa = 3.58FGPPKK242 pKa = 9.76EE243 pKa = 4.17KK244 pKa = 10.68SVTHH248 pKa = 7.09GITGGIDD255 pKa = 3.25LQALSNGTGAIWMGKK270 pKa = 8.83GPQGEE275 pKa = 4.69GPSSNITGAYY285 pKa = 9.27IIMLPGPGFNGPMISLEE302 pKa = 4.06TTNNHH307 pKa = 5.23RR308 pKa = 11.84VWVFRR313 pKa = 11.84DD314 pKa = 3.47PGRR317 pKa = 11.84NRR319 pKa = 11.84QGQSSRR325 pKa = 3.48
MM1 pKa = 7.49PRR3 pKa = 11.84SFMWAPDD10 pKa = 3.59PQAWLRR16 pKa = 11.84DD17 pKa = 3.89DD18 pKa = 4.78AVPTALAVLWAAMALEE34 pKa = 4.71GRR36 pKa = 11.84CGEE39 pKa = 4.42RR40 pKa = 11.84GATPARR46 pKa = 11.84RR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84QAPVPRR58 pKa = 11.84TRR60 pKa = 11.84LHH62 pKa = 6.47GLRR65 pKa = 11.84HH66 pKa = 4.37VCARR70 pKa = 11.84GSSIGRR76 pKa = 11.84RR77 pKa = 11.84TFWILALCTSLGLLLSWSSNRR98 pKa = 11.84LLHH101 pKa = 6.06WLSFPTYY108 pKa = 8.99TKK110 pKa = 10.87VHH112 pKa = 5.47TEE114 pKa = 3.7WAKK117 pKa = 10.13EE118 pKa = 3.91LPFPTVTICNNNPVRR133 pKa = 11.84FYY135 pKa = 11.58KK136 pKa = 9.61LTKK139 pKa = 9.75SDD141 pKa = 4.41LYY143 pKa = 11.08FAGHH147 pKa = 6.37WLGLLLANRR156 pKa = 11.84TARR159 pKa = 11.84PLVLDD164 pKa = 4.46LLQEE168 pKa = 4.02DD169 pKa = 3.71RR170 pKa = 11.84RR171 pKa = 11.84AWFHH175 pKa = 6.32KK176 pKa = 10.68LSDD179 pKa = 3.79FRR181 pKa = 11.84LFLPPRR187 pKa = 11.84HH188 pKa = 6.13FEE190 pKa = 4.03GTNLEE195 pKa = 4.16FMDD198 pKa = 4.46RR199 pKa = 11.84LSHH202 pKa = 6.68QLDD205 pKa = 4.24DD206 pKa = 4.07MLLSCKK212 pKa = 10.06YY213 pKa = 9.52RR214 pKa = 11.84GEE216 pKa = 4.02MCGPQNFSTATRR228 pKa = 11.84GRR230 pKa = 11.84VAAGNKK236 pKa = 9.94DD237 pKa = 3.58FGPPKK242 pKa = 9.76EE243 pKa = 4.17KK244 pKa = 10.68SVTHH248 pKa = 7.09GITGGIDD255 pKa = 3.25LQALSNGTGAIWMGKK270 pKa = 8.83GPQGEE275 pKa = 4.69GPSSNITGAYY285 pKa = 9.27IIMLPGPGFNGPMISLEE302 pKa = 4.06TTNNHH307 pKa = 5.23RR308 pKa = 11.84VWVFRR313 pKa = 11.84DD314 pKa = 3.47PGRR317 pKa = 11.84NRR319 pKa = 11.84QGQSSRR325 pKa = 3.48
Molecular weight: 36.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11792707 |
31 |
19435 |
487.3 |
54.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.942 ± 0.014 | 2.344 ± 0.014 |
5.011 ± 0.011 | 6.849 ± 0.019 |
3.625 ± 0.012 | 6.582 ± 0.02 |
2.612 ± 0.009 | 4.149 ± 0.014 |
5.368 ± 0.02 | 9.697 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.333 ± 0.007 | 3.535 ± 0.01 |
5.865 ± 0.022 | 4.611 ± 0.015 |
6.007 ± 0.014 | 8.538 ± 0.021 |
5.382 ± 0.013 | 6.555 ± 0.014 |
1.213 ± 0.006 | 2.702 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
