
Adlercreutzia equolifaciens subsp. celatus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Adlercreutzia; Adlercreutzia equolifaciens
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
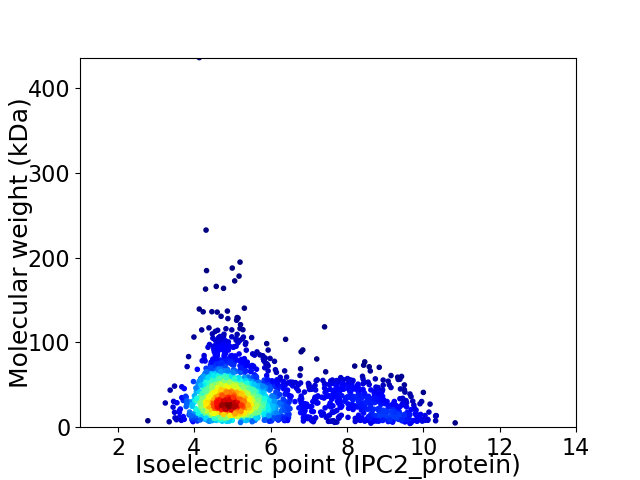
Virtual 2D-PAGE plot for 2195 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
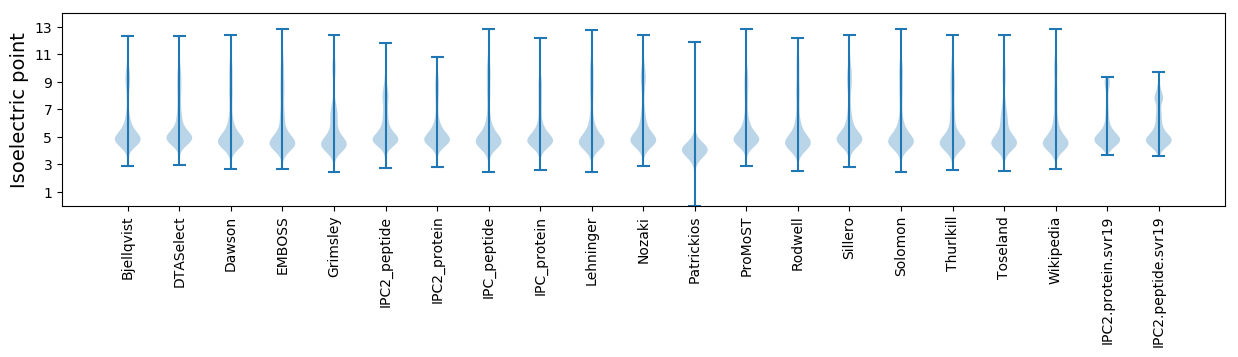
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369P163|A0A369P163_9ACTN Energy-dependent translational throttle protein EttA OS=Adlercreutzia equolifaciens subsp. celatus OX=394340 GN=ettA PE=3 SV=1
MM1 pKa = 7.47KK2 pKa = 10.19ASKK5 pKa = 10.41LIAAVVSGAAALALVGGIAYY25 pKa = 10.33AMEE28 pKa = 5.52PDD30 pKa = 4.49HH31 pKa = 7.49DD32 pKa = 4.58PATLPMPEE40 pKa = 4.27AAQDD44 pKa = 3.52TSIVKK49 pKa = 9.41QVASIIVYY57 pKa = 9.62EE58 pKa = 4.25DD59 pKa = 3.14QEE61 pKa = 4.39EE62 pKa = 4.29AVAMASEE69 pKa = 4.48FDD71 pKa = 4.93DD72 pKa = 3.6IHH74 pKa = 8.42DD75 pKa = 4.42EE76 pKa = 4.12LAMDD80 pKa = 4.69AVSSRR85 pKa = 11.84ASLSAKK91 pKa = 10.05HH92 pKa = 5.95SASHH96 pKa = 7.44DD97 pKa = 3.63ATAKK101 pKa = 8.83TAASAAGEE109 pKa = 4.31TPEE112 pKa = 5.48GSDD115 pKa = 4.54DD116 pKa = 4.53SIDD119 pKa = 3.67VEE121 pKa = 4.25AVDD124 pKa = 3.55VTEE127 pKa = 4.21PLEE130 pKa = 4.58TISPEE135 pKa = 3.91GDD137 pKa = 3.08EE138 pKa = 4.45MVADD142 pKa = 4.57SIDD145 pKa = 3.57VNGDD149 pKa = 2.44IVTYY153 pKa = 9.8VPSYY157 pKa = 10.65EE158 pKa = 4.33SASAPASGAGLWMGSDD174 pKa = 3.52STTDD178 pKa = 3.49GSWGYY183 pKa = 10.53FIGHH187 pKa = 6.35NPGDD191 pKa = 4.14FTCVMTLKK199 pKa = 10.81NGDD202 pKa = 3.95PVTVCDD208 pKa = 3.71SDD210 pKa = 4.65RR211 pKa = 11.84NTRR214 pKa = 11.84TYY216 pKa = 11.25SVVDD220 pKa = 3.56EE221 pKa = 5.64FIVPDD226 pKa = 3.44DD227 pKa = 4.65TYY229 pKa = 11.23WEE231 pKa = 5.31DD232 pKa = 3.12IQSRR236 pKa = 11.84VTDD239 pKa = 3.7YY240 pKa = 11.52GEE242 pKa = 4.65SIILQTCCGDD252 pKa = 3.33NAHH255 pKa = 6.06YY256 pKa = 10.03RR257 pKa = 11.84VVIAAA262 pKa = 4.3
MM1 pKa = 7.47KK2 pKa = 10.19ASKK5 pKa = 10.41LIAAVVSGAAALALVGGIAYY25 pKa = 10.33AMEE28 pKa = 5.52PDD30 pKa = 4.49HH31 pKa = 7.49DD32 pKa = 4.58PATLPMPEE40 pKa = 4.27AAQDD44 pKa = 3.52TSIVKK49 pKa = 9.41QVASIIVYY57 pKa = 9.62EE58 pKa = 4.25DD59 pKa = 3.14QEE61 pKa = 4.39EE62 pKa = 4.29AVAMASEE69 pKa = 4.48FDD71 pKa = 4.93DD72 pKa = 3.6IHH74 pKa = 8.42DD75 pKa = 4.42EE76 pKa = 4.12LAMDD80 pKa = 4.69AVSSRR85 pKa = 11.84ASLSAKK91 pKa = 10.05HH92 pKa = 5.95SASHH96 pKa = 7.44DD97 pKa = 3.63ATAKK101 pKa = 8.83TAASAAGEE109 pKa = 4.31TPEE112 pKa = 5.48GSDD115 pKa = 4.54DD116 pKa = 4.53SIDD119 pKa = 3.67VEE121 pKa = 4.25AVDD124 pKa = 3.55VTEE127 pKa = 4.21PLEE130 pKa = 4.58TISPEE135 pKa = 3.91GDD137 pKa = 3.08EE138 pKa = 4.45MVADD142 pKa = 4.57SIDD145 pKa = 3.57VNGDD149 pKa = 2.44IVTYY153 pKa = 9.8VPSYY157 pKa = 10.65EE158 pKa = 4.33SASAPASGAGLWMGSDD174 pKa = 3.52STTDD178 pKa = 3.49GSWGYY183 pKa = 10.53FIGHH187 pKa = 6.35NPGDD191 pKa = 4.14FTCVMTLKK199 pKa = 10.81NGDD202 pKa = 3.95PVTVCDD208 pKa = 3.71SDD210 pKa = 4.65RR211 pKa = 11.84NTRR214 pKa = 11.84TYY216 pKa = 11.25SVVDD220 pKa = 3.56EE221 pKa = 5.64FIVPDD226 pKa = 3.44DD227 pKa = 4.65TYY229 pKa = 11.23WEE231 pKa = 5.31DD232 pKa = 3.12IQSRR236 pKa = 11.84VTDD239 pKa = 3.7YY240 pKa = 11.52GEE242 pKa = 4.65SIILQTCCGDD252 pKa = 3.33NAHH255 pKa = 6.06YY256 pKa = 10.03RR257 pKa = 11.84VVIAAA262 pKa = 4.3
Molecular weight: 27.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369P1C8|A0A369P1C8_9ACTN Peptidase OS=Adlercreutzia equolifaciens subsp. celatus OX=394340 GN=C1850_03650 PE=4 SV=1
MM1 pKa = 7.25NVGSIDD7 pKa = 4.8LIPRR11 pKa = 11.84TLFHH15 pKa = 6.66GRR17 pKa = 11.84GTISCLPMNVYY28 pKa = 10.56LSHH31 pKa = 6.01NTALLFWRR39 pKa = 11.84AWSAAAAIPLCVFHH53 pKa = 7.53DD54 pKa = 4.45LGVADD59 pKa = 5.17ASLFPATLFPTSEE72 pKa = 4.07VLRR75 pKa = 11.84NEE77 pKa = 4.07AVLSRR82 pKa = 11.84DD83 pKa = 2.76VRR85 pKa = 11.84TILEE89 pKa = 4.51DD90 pKa = 3.58ALQSLAGAGAARR102 pKa = 11.84LADD105 pKa = 4.27AGSVASPDD113 pKa = 3.4GSAAPAAAVGGMLPQTTAVASALKK137 pKa = 10.15EE138 pKa = 4.16VVRR141 pKa = 11.84SRR143 pKa = 11.84GAAPLHH149 pKa = 5.9IMVAKK154 pKa = 10.39GHH156 pKa = 5.13GTRR159 pKa = 11.84NSSDD163 pKa = 4.27LIRR166 pKa = 11.84HH167 pKa = 6.16RR168 pKa = 11.84SVAAFPRR175 pKa = 11.84QAFIKK180 pKa = 8.81VAPGVFVCSPEE191 pKa = 4.04LVFVQMVSSLTRR203 pKa = 11.84GGLLAVGYY211 pKa = 6.94EE212 pKa = 4.11LCGCYY217 pKa = 9.84PLGEE221 pKa = 4.29EE222 pKa = 4.35RR223 pKa = 11.84VGALVRR229 pKa = 11.84RR230 pKa = 11.84PLSSPRR236 pKa = 11.84RR237 pKa = 11.84LTAFAARR244 pKa = 11.84AKK246 pKa = 10.24GLAGVKK252 pKa = 8.76VARR255 pKa = 11.84SAAKK259 pKa = 9.69QVLAKK264 pKa = 10.25SGSLMEE270 pKa = 4.4TEE272 pKa = 4.19VSIIAFSLFSQGGLGLRR289 pKa = 11.84TPLLNEE295 pKa = 3.73PVALSDD301 pKa = 3.81RR302 pKa = 11.84ARR304 pKa = 11.84TATGLSRR311 pKa = 11.84VVCDD315 pKa = 3.66WLWPAEE321 pKa = 4.09RR322 pKa = 11.84VVLEE326 pKa = 4.04YY327 pKa = 10.65DD328 pKa = 3.08GHH330 pKa = 6.87DD331 pKa = 3.37AHH333 pKa = 7.61SSPQQRR339 pKa = 11.84AHH341 pKa = 7.44DD342 pKa = 3.78ARR344 pKa = 11.84KK345 pKa = 9.72RR346 pKa = 11.84DD347 pKa = 3.54ALRR350 pKa = 11.84IDD352 pKa = 4.06GFDD355 pKa = 3.66LAVLTSSQFHH365 pKa = 6.71HH366 pKa = 7.0VNQCTEE372 pKa = 4.04LLRR375 pKa = 11.84GVARR379 pKa = 11.84SAGQRR384 pKa = 11.84SPALKK389 pKa = 10.41LEE391 pKa = 4.05HH392 pKa = 6.69LPRR395 pKa = 11.84HH396 pKa = 5.2LALRR400 pKa = 11.84EE401 pKa = 3.9QVRR404 pKa = 11.84KK405 pKa = 8.89HH406 pKa = 5.63HH407 pKa = 6.41RR408 pKa = 11.84AHH410 pKa = 6.15FPSRR414 pKa = 11.84FRR416 pKa = 11.84RR417 pKa = 11.84ARR419 pKa = 11.84SS420 pKa = 3.13
MM1 pKa = 7.25NVGSIDD7 pKa = 4.8LIPRR11 pKa = 11.84TLFHH15 pKa = 6.66GRR17 pKa = 11.84GTISCLPMNVYY28 pKa = 10.56LSHH31 pKa = 6.01NTALLFWRR39 pKa = 11.84AWSAAAAIPLCVFHH53 pKa = 7.53DD54 pKa = 4.45LGVADD59 pKa = 5.17ASLFPATLFPTSEE72 pKa = 4.07VLRR75 pKa = 11.84NEE77 pKa = 4.07AVLSRR82 pKa = 11.84DD83 pKa = 2.76VRR85 pKa = 11.84TILEE89 pKa = 4.51DD90 pKa = 3.58ALQSLAGAGAARR102 pKa = 11.84LADD105 pKa = 4.27AGSVASPDD113 pKa = 3.4GSAAPAAAVGGMLPQTTAVASALKK137 pKa = 10.15EE138 pKa = 4.16VVRR141 pKa = 11.84SRR143 pKa = 11.84GAAPLHH149 pKa = 5.9IMVAKK154 pKa = 10.39GHH156 pKa = 5.13GTRR159 pKa = 11.84NSSDD163 pKa = 4.27LIRR166 pKa = 11.84HH167 pKa = 6.16RR168 pKa = 11.84SVAAFPRR175 pKa = 11.84QAFIKK180 pKa = 8.81VAPGVFVCSPEE191 pKa = 4.04LVFVQMVSSLTRR203 pKa = 11.84GGLLAVGYY211 pKa = 6.94EE212 pKa = 4.11LCGCYY217 pKa = 9.84PLGEE221 pKa = 4.29EE222 pKa = 4.35RR223 pKa = 11.84VGALVRR229 pKa = 11.84RR230 pKa = 11.84PLSSPRR236 pKa = 11.84RR237 pKa = 11.84LTAFAARR244 pKa = 11.84AKK246 pKa = 10.24GLAGVKK252 pKa = 8.76VARR255 pKa = 11.84SAAKK259 pKa = 9.69QVLAKK264 pKa = 10.25SGSLMEE270 pKa = 4.4TEE272 pKa = 4.19VSIIAFSLFSQGGLGLRR289 pKa = 11.84TPLLNEE295 pKa = 3.73PVALSDD301 pKa = 3.81RR302 pKa = 11.84ARR304 pKa = 11.84TATGLSRR311 pKa = 11.84VVCDD315 pKa = 3.66WLWPAEE321 pKa = 4.09RR322 pKa = 11.84VVLEE326 pKa = 4.04YY327 pKa = 10.65DD328 pKa = 3.08GHH330 pKa = 6.87DD331 pKa = 3.37AHH333 pKa = 7.61SSPQQRR339 pKa = 11.84AHH341 pKa = 7.44DD342 pKa = 3.78ARR344 pKa = 11.84KK345 pKa = 9.72RR346 pKa = 11.84DD347 pKa = 3.54ALRR350 pKa = 11.84IDD352 pKa = 4.06GFDD355 pKa = 3.66LAVLTSSQFHH365 pKa = 6.71HH366 pKa = 7.0VNQCTEE372 pKa = 4.04LLRR375 pKa = 11.84GVARR379 pKa = 11.84SAGQRR384 pKa = 11.84SPALKK389 pKa = 10.41LEE391 pKa = 4.05HH392 pKa = 6.69LPRR395 pKa = 11.84HH396 pKa = 5.2LALRR400 pKa = 11.84EE401 pKa = 3.9QVRR404 pKa = 11.84KK405 pKa = 8.89HH406 pKa = 5.63HH407 pKa = 6.41RR408 pKa = 11.84AHH410 pKa = 6.15FPSRR414 pKa = 11.84FRR416 pKa = 11.84RR417 pKa = 11.84ARR419 pKa = 11.84SS420 pKa = 3.13
Molecular weight: 45.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
753156 |
37 |
4028 |
343.1 |
37.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.51 ± 0.079 | 1.599 ± 0.025 |
6.028 ± 0.053 | 6.899 ± 0.053 |
3.8 ± 0.037 | 8.278 ± 0.05 |
1.743 ± 0.022 | 4.917 ± 0.044 |
3.485 ± 0.04 | 9.326 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.647 ± 0.025 | 2.831 ± 0.033 |
4.528 ± 0.034 | 2.683 ± 0.023 |
5.96 ± 0.054 | 5.467 ± 0.045 |
5.32 ± 0.043 | 7.996 ± 0.041 |
1.16 ± 0.02 | 2.823 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
