
Rodentolepis nana (Dwarf tapeworm) (Hymenolepis nana)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Platyhelminthes; Cestoda; Eucestoda; Cyclophyllidea; Hymenolepididae; Rodentolepis
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
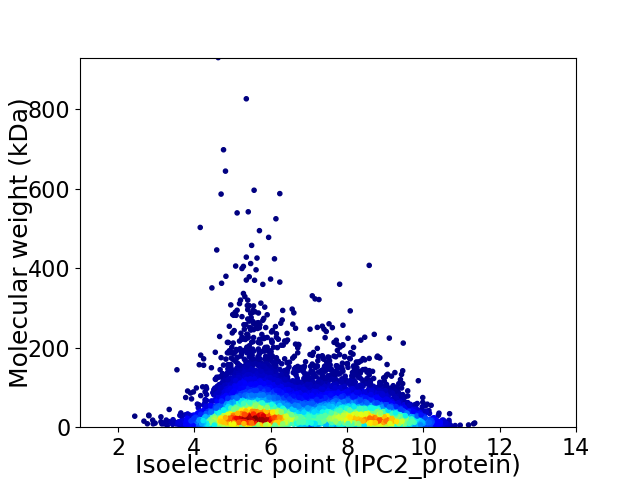
Virtual 2D-PAGE plot for 13592 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P7T2I2|A0A3P7T2I2_RODNA Uncharacterized protein OS=Rodentolepis nana OX=102285 GN=HNAJ_LOCUS7567 PE=4 SV=1
MM1 pKa = 7.54FGGTFSLPHH10 pKa = 5.95NFEE13 pKa = 3.88RR14 pKa = 11.84TAPIYY19 pKa = 10.79KK20 pKa = 10.24PGDD23 pKa = 3.42KK24 pKa = 10.38VLKK27 pKa = 9.76KK28 pKa = 10.39QEE30 pKa = 4.24LVAQSQADD38 pKa = 3.55SRR40 pKa = 11.84QQKK43 pKa = 9.0QAFFSNPQTEE53 pKa = 4.8LLCAMLEE60 pKa = 4.23LTNPNAVLLGQASYY74 pKa = 11.37NLSEE78 pKa = 4.33MANMLVEE85 pKa = 4.56RR86 pKa = 11.84GEE88 pKa = 4.54LDD90 pKa = 3.93EE91 pKa = 7.75DD92 pKa = 5.3DD93 pKa = 6.09DD94 pKa = 5.27NDD96 pKa = 4.12GCGVEE101 pKa = 4.29PSPAKK106 pKa = 10.2LARR109 pKa = 11.84VDD111 pKa = 4.2SNPEE115 pKa = 4.0EE116 pKa = 4.37INLDD120 pKa = 3.81DD121 pKa = 6.17LDD123 pKa = 4.67VDD125 pKa = 5.16DD126 pKa = 6.13EE127 pKa = 4.77EE128 pKa = 5.41EE129 pKa = 3.94NANNGGIVEE138 pKa = 4.61GYY140 pKa = 9.47EE141 pKa = 3.95PGVVPVNDD149 pKa = 2.89VDD151 pKa = 4.85YY152 pKa = 11.21NPQYY156 pKa = 11.09LAGCEE161 pKa = 4.01LPSASEE167 pKa = 4.28YY168 pKa = 10.09YY169 pKa = 10.1PEE171 pKa = 4.9GATVSAIEE179 pKa = 4.26VQYY182 pKa = 10.99SPNPIYY188 pKa = 10.42TSEE191 pKa = 4.04VSLTEE196 pKa = 4.03GNEE199 pKa = 4.01YY200 pKa = 10.81NPEE203 pKa = 4.09PLVKK207 pKa = 10.19NPEE210 pKa = 4.17VIDD213 pKa = 3.72IDD215 pKa = 4.71NLIDD219 pKa = 5.51DD220 pKa = 5.5DD221 pKa = 5.57DD222 pKa = 6.78DD223 pKa = 3.91EE224 pKa = 6.96ASADD228 pKa = 3.51NGPQEE233 pKa = 4.34EE234 pKa = 4.81YY235 pKa = 10.86EE236 pKa = 4.1PGGVKK241 pKa = 10.51LEE243 pKa = 4.15VSYY246 pKa = 11.26NPQPFIEE253 pKa = 4.76EE254 pKa = 4.35DD255 pKa = 3.58EE256 pKa = 4.5QPTKK260 pKa = 10.82QEE262 pKa = 4.54DD263 pKa = 3.18AGQDD267 pKa = 3.08EE268 pKa = 5.12GLTVV272 pKa = 3.33
MM1 pKa = 7.54FGGTFSLPHH10 pKa = 5.95NFEE13 pKa = 3.88RR14 pKa = 11.84TAPIYY19 pKa = 10.79KK20 pKa = 10.24PGDD23 pKa = 3.42KK24 pKa = 10.38VLKK27 pKa = 9.76KK28 pKa = 10.39QEE30 pKa = 4.24LVAQSQADD38 pKa = 3.55SRR40 pKa = 11.84QQKK43 pKa = 9.0QAFFSNPQTEE53 pKa = 4.8LLCAMLEE60 pKa = 4.23LTNPNAVLLGQASYY74 pKa = 11.37NLSEE78 pKa = 4.33MANMLVEE85 pKa = 4.56RR86 pKa = 11.84GEE88 pKa = 4.54LDD90 pKa = 3.93EE91 pKa = 7.75DD92 pKa = 5.3DD93 pKa = 6.09DD94 pKa = 5.27NDD96 pKa = 4.12GCGVEE101 pKa = 4.29PSPAKK106 pKa = 10.2LARR109 pKa = 11.84VDD111 pKa = 4.2SNPEE115 pKa = 4.0EE116 pKa = 4.37INLDD120 pKa = 3.81DD121 pKa = 6.17LDD123 pKa = 4.67VDD125 pKa = 5.16DD126 pKa = 6.13EE127 pKa = 4.77EE128 pKa = 5.41EE129 pKa = 3.94NANNGGIVEE138 pKa = 4.61GYY140 pKa = 9.47EE141 pKa = 3.95PGVVPVNDD149 pKa = 2.89VDD151 pKa = 4.85YY152 pKa = 11.21NPQYY156 pKa = 11.09LAGCEE161 pKa = 4.01LPSASEE167 pKa = 4.28YY168 pKa = 10.09YY169 pKa = 10.1PEE171 pKa = 4.9GATVSAIEE179 pKa = 4.26VQYY182 pKa = 10.99SPNPIYY188 pKa = 10.42TSEE191 pKa = 4.04VSLTEE196 pKa = 4.03GNEE199 pKa = 4.01YY200 pKa = 10.81NPEE203 pKa = 4.09PLVKK207 pKa = 10.19NPEE210 pKa = 4.17VIDD213 pKa = 3.72IDD215 pKa = 4.71NLIDD219 pKa = 5.51DD220 pKa = 5.5DD221 pKa = 5.57DD222 pKa = 6.78DD223 pKa = 3.91EE224 pKa = 6.96ASADD228 pKa = 3.51NGPQEE233 pKa = 4.34EE234 pKa = 4.81YY235 pKa = 10.86EE236 pKa = 4.1PGGVKK241 pKa = 10.51LEE243 pKa = 4.15VSYY246 pKa = 11.26NPQPFIEE253 pKa = 4.76EE254 pKa = 4.35DD255 pKa = 3.58EE256 pKa = 4.5QPTKK260 pKa = 10.82QEE262 pKa = 4.54DD263 pKa = 3.18AGQDD267 pKa = 3.08EE268 pKa = 5.12GLTVV272 pKa = 3.33
Molecular weight: 29.83 kDa
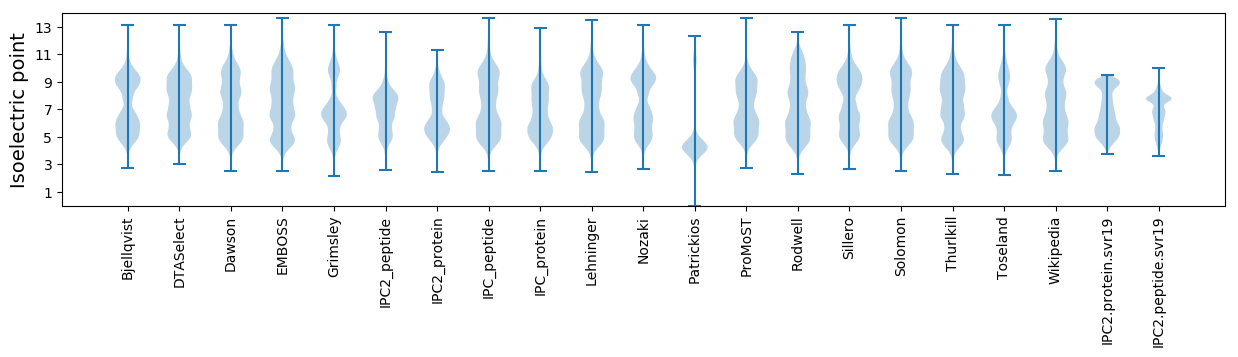
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A158QGF1|A0A158QGF1_RODNA Uncharacterized protein OS=Rodentolepis nana OX=102285 GN=HNAJ_LOCUS3 PE=3 SV=1
MM1 pKa = 7.2NLVRR5 pKa = 11.84RR6 pKa = 11.84NLVTLMRR13 pKa = 11.84RR14 pKa = 11.84IQMNLVTLMRR24 pKa = 11.84RR25 pKa = 11.84IQMNLVKK32 pKa = 10.48RR33 pKa = 11.84NLVTLMRR40 pKa = 11.84RR41 pKa = 11.84IQMNLVKK48 pKa = 10.48RR49 pKa = 11.84NLVTLMRR56 pKa = 11.84RR57 pKa = 11.84IQMNLVGRR65 pKa = 11.84IPTLMM70 pKa = 4.37
MM1 pKa = 7.2NLVRR5 pKa = 11.84RR6 pKa = 11.84NLVTLMRR13 pKa = 11.84RR14 pKa = 11.84IQMNLVTLMRR24 pKa = 11.84RR25 pKa = 11.84IQMNLVKK32 pKa = 10.48RR33 pKa = 11.84NLVTLMRR40 pKa = 11.84RR41 pKa = 11.84IQMNLVKK48 pKa = 10.48RR49 pKa = 11.84NLVTLMRR56 pKa = 11.84RR57 pKa = 11.84IQMNLVGRR65 pKa = 11.84IPTLMM70 pKa = 4.37
Molecular weight: 8.53 kDa
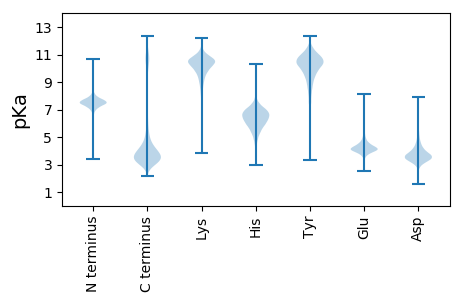
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5151847 |
29 |
8438 |
379.0 |
42.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.493 ± 0.019 | 2.026 ± 0.015 |
5.179 ± 0.017 | 6.359 ± 0.029 |
4.172 ± 0.016 | 5.499 ± 0.035 |
2.367 ± 0.009 | 5.645 ± 0.018 |
5.35 ± 0.021 | 9.557 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.008 | 4.757 ± 0.015 |
5.621 ± 0.026 | 4.037 ± 0.018 |
5.811 ± 0.019 | 9.449 ± 0.031 |
5.717 ± 0.019 | 5.915 ± 0.017 |
1.043 ± 0.007 | 2.819 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
