
Mycoplasma pneumoniae (strain ATCC 29342 / M129)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; Mycoplasma pneumoniae
Average proteome isoelectric point is 7.64
Get precalculated fractions of proteins
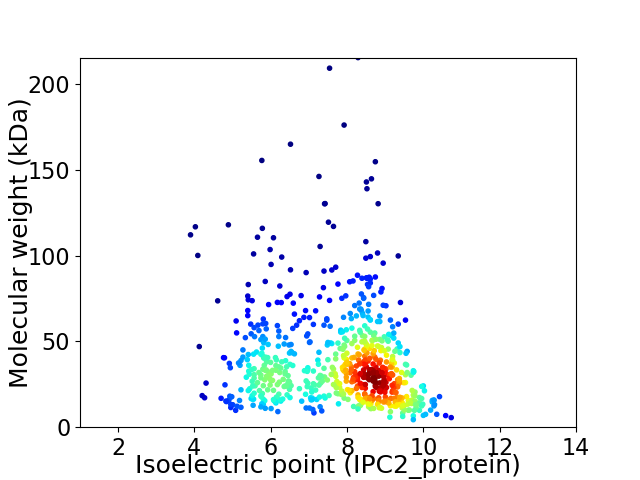
Virtual 2D-PAGE plot for 686 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P41204|RL16_MYCPN 50S ribosomal protein L16 OS=Mycoplasma pneumoniae (strain ATCC 29342 / M129) OX=272634 GN=rplP PE=3 SV=2
MM1 pKa = 8.28DD2 pKa = 4.51INKK5 pKa = 9.68PGWNQSDD12 pKa = 3.96QQATAYY18 pKa = 10.42DD19 pKa = 4.25PNQQQYY25 pKa = 9.74YY26 pKa = 9.97GDD28 pKa = 3.59GSTYY32 pKa = 10.44YY33 pKa = 10.98DD34 pKa = 4.02PDD36 pKa = 3.38QAVDD40 pKa = 3.85PNQAYY45 pKa = 10.16YY46 pKa = 9.83PDD48 pKa = 5.06PNTYY52 pKa = 9.85PDD54 pKa = 3.02AAAYY58 pKa = 9.46YY59 pKa = 10.51GYY61 pKa = 10.7GQDD64 pKa = 3.51GQAYY68 pKa = 7.49PQDD71 pKa = 3.95YY72 pKa = 10.84AQDD75 pKa = 4.0PNQAYY80 pKa = 9.86YY81 pKa = 11.06ADD83 pKa = 4.06PNAYY87 pKa = 9.52QDD89 pKa = 3.74PNAYY93 pKa = 8.03TDD95 pKa = 3.56PNAYY99 pKa = 9.83VDD101 pKa = 3.84PNAYY105 pKa = 9.55QDD107 pKa = 3.46PNAYY111 pKa = 9.92VDD113 pKa = 3.88PNNYY117 pKa = 8.3TDD119 pKa = 3.51PNAYY123 pKa = 9.75YY124 pKa = 10.62GYY126 pKa = 10.1GQDD129 pKa = 3.51GQAYY133 pKa = 7.49PQDD136 pKa = 3.95YY137 pKa = 10.84AQDD140 pKa = 4.0PNQAYY145 pKa = 9.86YY146 pKa = 11.06ADD148 pKa = 4.06PNAYY152 pKa = 9.52QDD154 pKa = 3.74PNAYY158 pKa = 8.12TDD160 pKa = 3.81PYY162 pKa = 10.31YY163 pKa = 10.33VTSTDD168 pKa = 3.03PNAYY172 pKa = 9.53YY173 pKa = 10.61GQVDD177 pKa = 3.96NVPALEE183 pKa = 4.56ASDD186 pKa = 3.96LAYY189 pKa = 10.17EE190 pKa = 4.23VTPQEE195 pKa = 3.84QAAEE199 pKa = 4.04QEE201 pKa = 4.62LFSEE205 pKa = 4.94PEE207 pKa = 3.9TKK209 pKa = 10.3VIRR212 pKa = 11.84EE213 pKa = 3.55IHH215 pKa = 5.98EE216 pKa = 4.35FPFEE220 pKa = 4.37KK221 pKa = 9.86IRR223 pKa = 11.84SYY225 pKa = 11.0FQTDD229 pKa = 3.44FDD231 pKa = 4.97SYY233 pKa = 11.54NSRR236 pKa = 11.84LTQLKK241 pKa = 10.41DD242 pKa = 3.33KK243 pKa = 10.5LDD245 pKa = 3.6NAIFSMRR252 pKa = 11.84KK253 pKa = 9.92AIDD256 pKa = 3.56TVKK259 pKa = 10.6EE260 pKa = 3.78NSANLQIMKK269 pKa = 10.51QNFEE273 pKa = 4.21RR274 pKa = 11.84QLKK277 pKa = 8.02EE278 pKa = 3.67QQTQRR283 pKa = 11.84LTSNTDD289 pKa = 2.94AEE291 pKa = 4.65KK292 pKa = 10.47IGAKK296 pKa = 9.68INQLEE301 pKa = 4.05EE302 pKa = 3.76RR303 pKa = 11.84MQRR306 pKa = 11.84LSRR309 pKa = 11.84TMEE312 pKa = 3.98SVEE315 pKa = 3.94WTKK318 pKa = 11.02KK319 pKa = 9.23EE320 pKa = 3.77PRR322 pKa = 11.84QEE324 pKa = 3.95QFDD327 pKa = 3.94PRR329 pKa = 11.84FVDD332 pKa = 3.54PRR334 pKa = 11.84NFNNYY339 pKa = 8.19VNNTDD344 pKa = 2.94TMMSMFEE351 pKa = 4.31KK352 pKa = 10.53VLMMNLLRR360 pKa = 11.84STTPVQPPVQYY371 pKa = 7.39FTPQPLTASPRR382 pKa = 11.84PVYY385 pKa = 9.98EE386 pKa = 4.1EE387 pKa = 5.06PISASFRR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84GYY398 pKa = 10.58RR399 pKa = 11.84GDD401 pKa = 3.52EE402 pKa = 4.35FYY404 pKa = 11.05EE405 pKa = 4.12
MM1 pKa = 8.28DD2 pKa = 4.51INKK5 pKa = 9.68PGWNQSDD12 pKa = 3.96QQATAYY18 pKa = 10.42DD19 pKa = 4.25PNQQQYY25 pKa = 9.74YY26 pKa = 9.97GDD28 pKa = 3.59GSTYY32 pKa = 10.44YY33 pKa = 10.98DD34 pKa = 4.02PDD36 pKa = 3.38QAVDD40 pKa = 3.85PNQAYY45 pKa = 10.16YY46 pKa = 9.83PDD48 pKa = 5.06PNTYY52 pKa = 9.85PDD54 pKa = 3.02AAAYY58 pKa = 9.46YY59 pKa = 10.51GYY61 pKa = 10.7GQDD64 pKa = 3.51GQAYY68 pKa = 7.49PQDD71 pKa = 3.95YY72 pKa = 10.84AQDD75 pKa = 4.0PNQAYY80 pKa = 9.86YY81 pKa = 11.06ADD83 pKa = 4.06PNAYY87 pKa = 9.52QDD89 pKa = 3.74PNAYY93 pKa = 8.03TDD95 pKa = 3.56PNAYY99 pKa = 9.83VDD101 pKa = 3.84PNAYY105 pKa = 9.55QDD107 pKa = 3.46PNAYY111 pKa = 9.92VDD113 pKa = 3.88PNNYY117 pKa = 8.3TDD119 pKa = 3.51PNAYY123 pKa = 9.75YY124 pKa = 10.62GYY126 pKa = 10.1GQDD129 pKa = 3.51GQAYY133 pKa = 7.49PQDD136 pKa = 3.95YY137 pKa = 10.84AQDD140 pKa = 4.0PNQAYY145 pKa = 9.86YY146 pKa = 11.06ADD148 pKa = 4.06PNAYY152 pKa = 9.52QDD154 pKa = 3.74PNAYY158 pKa = 8.12TDD160 pKa = 3.81PYY162 pKa = 10.31YY163 pKa = 10.33VTSTDD168 pKa = 3.03PNAYY172 pKa = 9.53YY173 pKa = 10.61GQVDD177 pKa = 3.96NVPALEE183 pKa = 4.56ASDD186 pKa = 3.96LAYY189 pKa = 10.17EE190 pKa = 4.23VTPQEE195 pKa = 3.84QAAEE199 pKa = 4.04QEE201 pKa = 4.62LFSEE205 pKa = 4.94PEE207 pKa = 3.9TKK209 pKa = 10.3VIRR212 pKa = 11.84EE213 pKa = 3.55IHH215 pKa = 5.98EE216 pKa = 4.35FPFEE220 pKa = 4.37KK221 pKa = 9.86IRR223 pKa = 11.84SYY225 pKa = 11.0FQTDD229 pKa = 3.44FDD231 pKa = 4.97SYY233 pKa = 11.54NSRR236 pKa = 11.84LTQLKK241 pKa = 10.41DD242 pKa = 3.33KK243 pKa = 10.5LDD245 pKa = 3.6NAIFSMRR252 pKa = 11.84KK253 pKa = 9.92AIDD256 pKa = 3.56TVKK259 pKa = 10.6EE260 pKa = 3.78NSANLQIMKK269 pKa = 10.51QNFEE273 pKa = 4.21RR274 pKa = 11.84QLKK277 pKa = 8.02EE278 pKa = 3.67QQTQRR283 pKa = 11.84LTSNTDD289 pKa = 2.94AEE291 pKa = 4.65KK292 pKa = 10.47IGAKK296 pKa = 9.68INQLEE301 pKa = 4.05EE302 pKa = 3.76RR303 pKa = 11.84MQRR306 pKa = 11.84LSRR309 pKa = 11.84TMEE312 pKa = 3.98SVEE315 pKa = 3.94WTKK318 pKa = 11.02KK319 pKa = 9.23EE320 pKa = 3.77PRR322 pKa = 11.84QEE324 pKa = 3.95QFDD327 pKa = 3.94PRR329 pKa = 11.84FVDD332 pKa = 3.54PRR334 pKa = 11.84NFNNYY339 pKa = 8.19VNNTDD344 pKa = 2.94TMMSMFEE351 pKa = 4.31KK352 pKa = 10.53VLMMNLLRR360 pKa = 11.84STTPVQPPVQYY371 pKa = 7.39FTPQPLTASPRR382 pKa = 11.84PVYY385 pKa = 9.98EE386 pKa = 4.1EE387 pKa = 5.06PISASFRR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84GYY398 pKa = 10.58RR399 pKa = 11.84GDD401 pKa = 3.52EE402 pKa = 4.35FYY404 pKa = 11.05EE405 pKa = 4.12
Molecular weight: 47.04 kDa
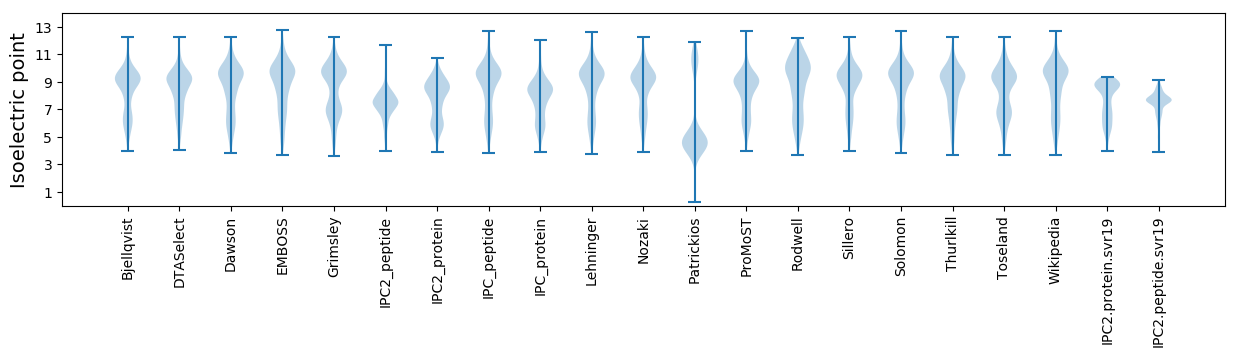
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P75413|Y368_MYCPN UPF0134 protein MPN_368 OS=Mycoplasma pneumoniae (strain ATCC 29342 / M129) OX=272634 GN=MPN_368 PE=3 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84IEE4 pKa = 4.24AANLAGSLWICSVVNHH20 pKa = 6.38GVQGVGVPSWVPDD33 pKa = 3.89PEE35 pKa = 5.15LEE37 pKa = 4.19GAVPKK42 pKa = 10.74SSALSWTCWLLLEE55 pKa = 4.69PRR57 pKa = 11.84LIGALARR64 pKa = 11.84LLVSSSIWPLSSEE77 pKa = 3.79SDD79 pKa = 3.65FFFTATCNALTLVSPDD95 pKa = 3.6EE96 pKa = 4.13PHH98 pKa = 6.61VGWIGQIQMWLKK110 pKa = 9.68NQWPQRR116 pKa = 11.84PGVFHH121 pKa = 7.02CSSRR125 pKa = 11.84CPPRR129 pKa = 11.84RR130 pKa = 11.84SSPSSQTLPRR140 pKa = 11.84WWKK143 pKa = 9.92YY144 pKa = 9.76FDD146 pKa = 3.49HH147 pKa = 6.83SRR149 pKa = 11.84FAAVVSPTPFATAHH163 pKa = 5.0STPRR167 pKa = 11.84CAARR171 pKa = 11.84VKK173 pKa = 10.41RR174 pKa = 11.84QTGRR178 pKa = 11.84DD179 pKa = 3.3WRR181 pKa = 11.84GLAPPRR187 pKa = 11.84RR188 pKa = 11.84GPCFWPRR195 pKa = 11.84FTVGVQPHH203 pKa = 6.43SNQSRR208 pKa = 11.84QRR210 pKa = 11.84GG211 pKa = 3.49
MM1 pKa = 7.73RR2 pKa = 11.84IEE4 pKa = 4.24AANLAGSLWICSVVNHH20 pKa = 6.38GVQGVGVPSWVPDD33 pKa = 3.89PEE35 pKa = 5.15LEE37 pKa = 4.19GAVPKK42 pKa = 10.74SSALSWTCWLLLEE55 pKa = 4.69PRR57 pKa = 11.84LIGALARR64 pKa = 11.84LLVSSSIWPLSSEE77 pKa = 3.79SDD79 pKa = 3.65FFFTATCNALTLVSPDD95 pKa = 3.6EE96 pKa = 4.13PHH98 pKa = 6.61VGWIGQIQMWLKK110 pKa = 9.68NQWPQRR116 pKa = 11.84PGVFHH121 pKa = 7.02CSSRR125 pKa = 11.84CPPRR129 pKa = 11.84RR130 pKa = 11.84SSPSSQTLPRR140 pKa = 11.84WWKK143 pKa = 9.92YY144 pKa = 9.76FDD146 pKa = 3.49HH147 pKa = 6.83SRR149 pKa = 11.84FAAVVSPTPFATAHH163 pKa = 5.0STPRR167 pKa = 11.84CAARR171 pKa = 11.84VKK173 pKa = 10.41RR174 pKa = 11.84QTGRR178 pKa = 11.84DD179 pKa = 3.3WRR181 pKa = 11.84GLAPPRR187 pKa = 11.84RR188 pKa = 11.84GPCFWPRR195 pKa = 11.84FTVGVQPHH203 pKa = 6.43SNQSRR208 pKa = 11.84QRR210 pKa = 11.84GG211 pKa = 3.49
Molecular weight: 23.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
239873 |
37 |
1882 |
349.7 |
39.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.672 ± 0.08 | 0.75 ± 0.027 |
4.974 ± 0.072 | 5.681 ± 0.12 |
5.599 ± 0.088 | 5.522 ± 0.105 |
1.81 ± 0.043 | 6.627 ± 0.094 |
8.562 ± 0.103 | 10.35 ± 0.112 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.591 ± 0.034 | 6.209 ± 0.067 |
3.49 ± 0.076 | 5.372 ± 0.104 |
3.487 ± 0.057 | 6.462 ± 0.094 |
5.956 ± 0.073 | 6.479 ± 0.077 |
1.18 ± 0.037 | 3.228 ± 0.056 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
