
Candidatus Cyclonatronum proteinivorum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; unclassified Balneolaeota; Candidatus Cyclonatronum
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
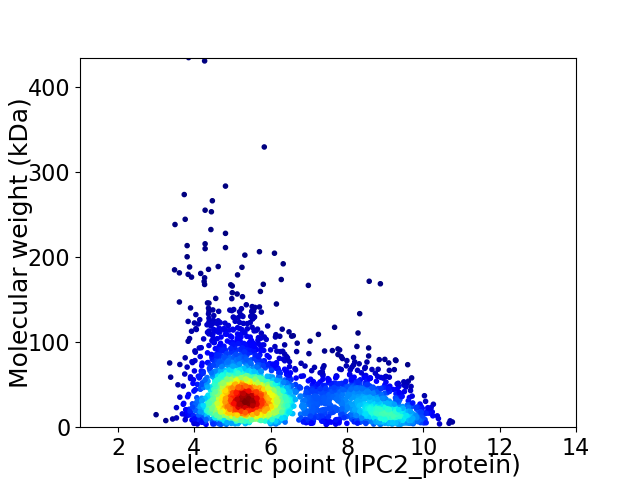
Virtual 2D-PAGE plot for 3251 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345UJA3|A0A345UJA3_9BACT His Kinase A (Phospho-acceptor) domain-containing protein OS=Candidatus Cyclonatronum proteinivorum OX=1457365 GN=CYPRO_1298 PE=4 SV=1
MM1 pKa = 8.19LFMLLLWQVIPMSAVQANPPSCWGEE26 pKa = 4.25GNCDD30 pKa = 2.87MSRR33 pKa = 11.84PYY35 pKa = 10.34FQLNIDD41 pKa = 3.46KK42 pKa = 10.97SNYY45 pKa = 8.69VLPNGMEE52 pKa = 4.16YY53 pKa = 10.79NQVLNMQHH61 pKa = 6.82SNTKK65 pKa = 10.5SPDD68 pKa = 3.18LTFQSASTLGLTGATQIGQMAVDD91 pKa = 3.73AQGNRR96 pKa = 11.84YY97 pKa = 6.86VTGGFTGTISYY108 pKa = 10.65DD109 pKa = 3.43GVIVEE114 pKa = 4.49STGGYY119 pKa = 10.53DD120 pKa = 3.19VFIAKK125 pKa = 9.28FSPEE129 pKa = 4.07GTLLWYY135 pKa = 10.32QLAHH139 pKa = 6.22GSEE142 pKa = 4.31EE143 pKa = 4.17VEE145 pKa = 4.56EE146 pKa = 4.22EE147 pKa = 4.03FSLDD151 pKa = 3.23GGLTLAVDD159 pKa = 3.73DD160 pKa = 5.01HH161 pKa = 7.32GNVYY165 pKa = 10.72VGGAFVKK172 pKa = 10.16EE173 pKa = 3.74LHH175 pKa = 5.92FTDD178 pKa = 3.62NNGEE182 pKa = 4.17ILSSLSDD189 pKa = 3.52GRR191 pKa = 11.84DD192 pKa = 3.18DD193 pKa = 5.72DD194 pKa = 5.82LINLEE199 pKa = 4.59LFVAKK204 pKa = 10.53YY205 pKa = 7.72DD206 pKa = 3.94TNGNFQWALGGDD218 pKa = 4.02SGSEE222 pKa = 3.68ASPNSLEE229 pKa = 4.79LGINSVNSVMIDD241 pKa = 3.24TDD243 pKa = 4.16GYY245 pKa = 9.34PYY247 pKa = 10.76VAGGFSGTNLFGEE260 pKa = 4.65EE261 pKa = 3.91VTVQGEE267 pKa = 4.27SDD269 pKa = 3.7FFIASLDD276 pKa = 3.33KK277 pKa = 11.22DD278 pKa = 3.8GNYY281 pKa = 10.36LYY283 pKa = 9.74WANVFGTPGRR293 pKa = 11.84DD294 pKa = 3.08LASSISVDD302 pKa = 2.84ALGYY306 pKa = 10.79LNILGIVGEE315 pKa = 4.26GRR317 pKa = 11.84MYY319 pKa = 11.13LPDD322 pKa = 4.42SDD324 pKa = 5.41IYY326 pKa = 10.52WDD328 pKa = 3.96NDD330 pKa = 3.16TGNADD335 pKa = 3.86TFVISYY341 pKa = 9.76DD342 pKa = 4.05VNGEE346 pKa = 3.85WYY348 pKa = 9.51FASFMGAGDD357 pKa = 4.75DD358 pKa = 3.5IVGKK362 pKa = 9.89SVVSAADD369 pKa = 3.4GSFYY373 pKa = 11.17VGGFFSGTDD382 pKa = 3.25AYY384 pKa = 11.17FEE386 pKa = 5.08GYY388 pKa = 10.68NKK390 pKa = 9.66TFDD393 pKa = 4.06AMGTEE398 pKa = 4.37DD399 pKa = 4.85AFLVKK404 pKa = 10.42YY405 pKa = 10.59DD406 pKa = 3.89IDD408 pKa = 5.28GDD410 pKa = 4.57AIWVRR415 pKa = 11.84QFGFDD420 pKa = 3.24RR421 pKa = 11.84ATVDD425 pKa = 4.43VVTLDD430 pKa = 3.26EE431 pKa = 4.49NEE433 pKa = 4.09NVFVLGRR440 pKa = 11.84YY441 pKa = 8.95SDD443 pKa = 4.83SIIFDD448 pKa = 3.66IDD450 pKa = 3.29SDD452 pKa = 4.42SPVILTTEE460 pKa = 4.45SINNIYY466 pKa = 8.53MAKK469 pKa = 9.82FDD471 pKa = 3.85EE472 pKa = 4.74SGNFIWAKK480 pKa = 10.21NIEE483 pKa = 4.24GSGAEE488 pKa = 3.97SRR490 pKa = 11.84EE491 pKa = 3.91LVFDD495 pKa = 3.85RR496 pKa = 11.84EE497 pKa = 4.26TRR499 pKa = 11.84PFGTNPLDD507 pKa = 4.63IITSAYY513 pKa = 9.99NGGEE517 pKa = 4.03IILAGDD523 pKa = 3.97FDD525 pKa = 4.37GTLQLDD531 pKa = 4.96DD532 pKa = 3.89ITLTSDD538 pKa = 3.56GTRR541 pKa = 11.84KK542 pKa = 9.72IFMAVMPTGGAVSVGEE558 pKa = 4.7DD559 pKa = 3.42DD560 pKa = 5.24HH561 pKa = 7.01NIPTGIQLSQNYY573 pKa = 7.57PNPFNPTTNISFNLPQASQVSITVYY598 pKa = 9.28TVMGQRR604 pKa = 11.84VGTISNDD611 pKa = 2.66MFAAGQHH618 pKa = 4.57TVTWDD623 pKa = 3.2AAGLSSGTYY632 pKa = 8.75MYY634 pKa = 10.86RR635 pKa = 11.84LSADD639 pKa = 3.2NFSATRR645 pKa = 11.84KK646 pKa = 6.39MTLLKK651 pKa = 10.81
MM1 pKa = 8.19LFMLLLWQVIPMSAVQANPPSCWGEE26 pKa = 4.25GNCDD30 pKa = 2.87MSRR33 pKa = 11.84PYY35 pKa = 10.34FQLNIDD41 pKa = 3.46KK42 pKa = 10.97SNYY45 pKa = 8.69VLPNGMEE52 pKa = 4.16YY53 pKa = 10.79NQVLNMQHH61 pKa = 6.82SNTKK65 pKa = 10.5SPDD68 pKa = 3.18LTFQSASTLGLTGATQIGQMAVDD91 pKa = 3.73AQGNRR96 pKa = 11.84YY97 pKa = 6.86VTGGFTGTISYY108 pKa = 10.65DD109 pKa = 3.43GVIVEE114 pKa = 4.49STGGYY119 pKa = 10.53DD120 pKa = 3.19VFIAKK125 pKa = 9.28FSPEE129 pKa = 4.07GTLLWYY135 pKa = 10.32QLAHH139 pKa = 6.22GSEE142 pKa = 4.31EE143 pKa = 4.17VEE145 pKa = 4.56EE146 pKa = 4.22EE147 pKa = 4.03FSLDD151 pKa = 3.23GGLTLAVDD159 pKa = 3.73DD160 pKa = 5.01HH161 pKa = 7.32GNVYY165 pKa = 10.72VGGAFVKK172 pKa = 10.16EE173 pKa = 3.74LHH175 pKa = 5.92FTDD178 pKa = 3.62NNGEE182 pKa = 4.17ILSSLSDD189 pKa = 3.52GRR191 pKa = 11.84DD192 pKa = 3.18DD193 pKa = 5.72DD194 pKa = 5.82LINLEE199 pKa = 4.59LFVAKK204 pKa = 10.53YY205 pKa = 7.72DD206 pKa = 3.94TNGNFQWALGGDD218 pKa = 4.02SGSEE222 pKa = 3.68ASPNSLEE229 pKa = 4.79LGINSVNSVMIDD241 pKa = 3.24TDD243 pKa = 4.16GYY245 pKa = 9.34PYY247 pKa = 10.76VAGGFSGTNLFGEE260 pKa = 4.65EE261 pKa = 3.91VTVQGEE267 pKa = 4.27SDD269 pKa = 3.7FFIASLDD276 pKa = 3.33KK277 pKa = 11.22DD278 pKa = 3.8GNYY281 pKa = 10.36LYY283 pKa = 9.74WANVFGTPGRR293 pKa = 11.84DD294 pKa = 3.08LASSISVDD302 pKa = 2.84ALGYY306 pKa = 10.79LNILGIVGEE315 pKa = 4.26GRR317 pKa = 11.84MYY319 pKa = 11.13LPDD322 pKa = 4.42SDD324 pKa = 5.41IYY326 pKa = 10.52WDD328 pKa = 3.96NDD330 pKa = 3.16TGNADD335 pKa = 3.86TFVISYY341 pKa = 9.76DD342 pKa = 4.05VNGEE346 pKa = 3.85WYY348 pKa = 9.51FASFMGAGDD357 pKa = 4.75DD358 pKa = 3.5IVGKK362 pKa = 9.89SVVSAADD369 pKa = 3.4GSFYY373 pKa = 11.17VGGFFSGTDD382 pKa = 3.25AYY384 pKa = 11.17FEE386 pKa = 5.08GYY388 pKa = 10.68NKK390 pKa = 9.66TFDD393 pKa = 4.06AMGTEE398 pKa = 4.37DD399 pKa = 4.85AFLVKK404 pKa = 10.42YY405 pKa = 10.59DD406 pKa = 3.89IDD408 pKa = 5.28GDD410 pKa = 4.57AIWVRR415 pKa = 11.84QFGFDD420 pKa = 3.24RR421 pKa = 11.84ATVDD425 pKa = 4.43VVTLDD430 pKa = 3.26EE431 pKa = 4.49NEE433 pKa = 4.09NVFVLGRR440 pKa = 11.84YY441 pKa = 8.95SDD443 pKa = 4.83SIIFDD448 pKa = 3.66IDD450 pKa = 3.29SDD452 pKa = 4.42SPVILTTEE460 pKa = 4.45SINNIYY466 pKa = 8.53MAKK469 pKa = 9.82FDD471 pKa = 3.85EE472 pKa = 4.74SGNFIWAKK480 pKa = 10.21NIEE483 pKa = 4.24GSGAEE488 pKa = 3.97SRR490 pKa = 11.84EE491 pKa = 3.91LVFDD495 pKa = 3.85RR496 pKa = 11.84EE497 pKa = 4.26TRR499 pKa = 11.84PFGTNPLDD507 pKa = 4.63IITSAYY513 pKa = 9.99NGGEE517 pKa = 4.03IILAGDD523 pKa = 3.97FDD525 pKa = 4.37GTLQLDD531 pKa = 4.96DD532 pKa = 3.89ITLTSDD538 pKa = 3.56GTRR541 pKa = 11.84KK542 pKa = 9.72IFMAVMPTGGAVSVGEE558 pKa = 4.7DD559 pKa = 3.42DD560 pKa = 5.24HH561 pKa = 7.01NIPTGIQLSQNYY573 pKa = 7.57PNPFNPTTNISFNLPQASQVSITVYY598 pKa = 9.28TVMGQRR604 pKa = 11.84VGTISNDD611 pKa = 2.66MFAAGQHH618 pKa = 4.57TVTWDD623 pKa = 3.2AAGLSSGTYY632 pKa = 8.75MYY634 pKa = 10.86RR635 pKa = 11.84LSADD639 pKa = 3.2NFSATRR645 pKa = 11.84KK646 pKa = 6.39MTLLKK651 pKa = 10.81
Molecular weight: 70.69 kDa
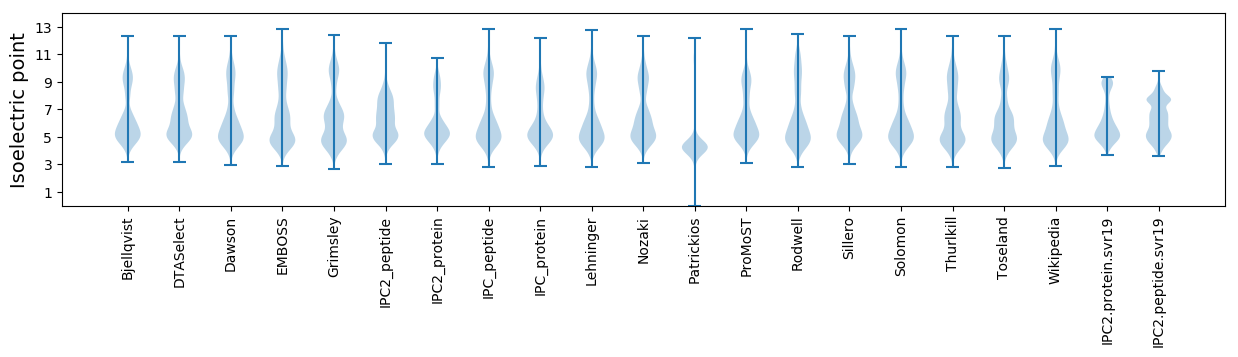
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345UL86|A0A345UL86_9BACT Uncharacterized protein OS=Candidatus Cyclonatronum proteinivorum OX=1457365 GN=CYPRO_1988 PE=4 SV=1
MM1 pKa = 7.59LPLQGSRR8 pKa = 11.84VFRR11 pKa = 11.84IGHH14 pKa = 5.58PRR16 pKa = 11.84RR17 pKa = 11.84DD18 pKa = 3.37AKK20 pKa = 11.1SRR22 pKa = 11.84VSTVPGARR30 pKa = 11.84TNLRR34 pKa = 11.84PTTGCIPHH42 pKa = 7.52KK43 pKa = 10.14SRR45 pKa = 11.84KK46 pKa = 7.84IPKK49 pKa = 9.3IRR51 pKa = 11.84VLSSRR56 pKa = 11.84TLTLPCVASGAEE68 pKa = 4.2LPHH71 pKa = 6.9PGYY74 pKa = 10.59HH75 pKa = 6.92IPPQTNKK82 pKa = 9.11IRR84 pKa = 11.84HH85 pKa = 4.93PTYY88 pKa = 10.44AVTTINHH95 pKa = 5.09EE96 pKa = 4.36TVIPEE101 pKa = 4.09TLKK104 pKa = 10.34PKK106 pKa = 10.52QIITSKK112 pKa = 8.18HH113 pKa = 4.75TGVIRR118 pKa = 11.84DD119 pKa = 4.57LPTSACDD126 pKa = 4.24LPCSPPPKK134 pKa = 9.88RR135 pKa = 11.84CTRR138 pKa = 11.84LCRR141 pKa = 11.84TDD143 pKa = 2.95IAAGNSYY150 pKa = 10.76CGISRR155 pKa = 11.84LADD158 pKa = 3.73PGG160 pKa = 3.66
MM1 pKa = 7.59LPLQGSRR8 pKa = 11.84VFRR11 pKa = 11.84IGHH14 pKa = 5.58PRR16 pKa = 11.84RR17 pKa = 11.84DD18 pKa = 3.37AKK20 pKa = 11.1SRR22 pKa = 11.84VSTVPGARR30 pKa = 11.84TNLRR34 pKa = 11.84PTTGCIPHH42 pKa = 7.52KK43 pKa = 10.14SRR45 pKa = 11.84KK46 pKa = 7.84IPKK49 pKa = 9.3IRR51 pKa = 11.84VLSSRR56 pKa = 11.84TLTLPCVASGAEE68 pKa = 4.2LPHH71 pKa = 6.9PGYY74 pKa = 10.59HH75 pKa = 6.92IPPQTNKK82 pKa = 9.11IRR84 pKa = 11.84HH85 pKa = 4.93PTYY88 pKa = 10.44AVTTINHH95 pKa = 5.09EE96 pKa = 4.36TVIPEE101 pKa = 4.09TLKK104 pKa = 10.34PKK106 pKa = 10.52QIITSKK112 pKa = 8.18HH113 pKa = 4.75TGVIRR118 pKa = 11.84DD119 pKa = 4.57LPTSACDD126 pKa = 4.24LPCSPPPKK134 pKa = 9.88RR135 pKa = 11.84CTRR138 pKa = 11.84LCRR141 pKa = 11.84TDD143 pKa = 2.95IAAGNSYY150 pKa = 10.76CGISRR155 pKa = 11.84LADD158 pKa = 3.73PGG160 pKa = 3.66
Molecular weight: 17.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1202591 |
29 |
4053 |
369.9 |
41.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.474 ± 0.042 | 0.684 ± 0.013 |
5.58 ± 0.03 | 6.714 ± 0.039 |
5.085 ± 0.036 | 7.398 ± 0.043 |
2.075 ± 0.024 | 6.095 ± 0.038 |
4.024 ± 0.062 | 9.942 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.418 ± 0.022 | 4.184 ± 0.039 |
4.616 ± 0.035 | 3.909 ± 0.031 |
5.93 ± 0.042 | 6.344 ± 0.03 |
5.713 ± 0.034 | 6.461 ± 0.032 |
1.247 ± 0.018 | 3.107 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
