
Sphingobacterium olei
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Sphingobacterium
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
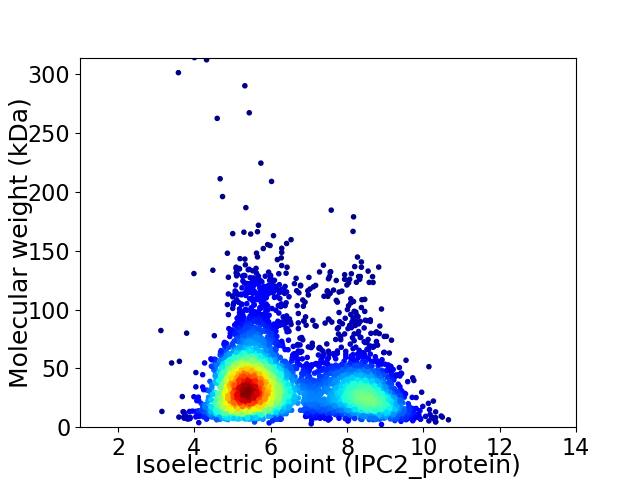
Virtual 2D-PAGE plot for 4341 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U0NKD8|A0A4U0NKD8_9SPHI Uncharacterized protein OS=Sphingobacterium olei OX=2571155 GN=FAZ15_15115 PE=4 SV=1
AA1 pKa = 7.03GAVYY5 pKa = 10.26NVAVAKK11 pKa = 10.87GKK13 pKa = 10.1DD14 pKa = 3.32PKK16 pKa = 11.17DD17 pKa = 3.78RR18 pKa = 11.84DD19 pKa = 3.87VEE21 pKa = 4.51GSSTDD26 pKa = 3.66NNPLDD31 pKa = 4.25PTDD34 pKa = 4.47PDD36 pKa = 4.03NPPVDD41 pKa = 4.38PNCPEE46 pKa = 4.03CTIVEE51 pKa = 4.16IDD53 pKa = 3.38QEE55 pKa = 4.45GAIEE59 pKa = 4.32VIKK62 pKa = 9.57TADD65 pKa = 2.9LSQRR69 pKa = 11.84FRR71 pKa = 11.84YY72 pKa = 10.04AGEE75 pKa = 3.85QIAYY79 pKa = 8.48EE80 pKa = 3.96ILIKK84 pKa = 10.95NVGNVTLHH92 pKa = 6.94DD93 pKa = 4.0IKK95 pKa = 11.07VTDD98 pKa = 4.36ANADD102 pKa = 3.66DD103 pKa = 4.41VNVGNIANLAVGEE116 pKa = 4.46SVTLQAYY123 pKa = 7.37HH124 pKa = 7.49TITQADD130 pKa = 3.86MEE132 pKa = 4.55RR133 pKa = 11.84GYY135 pKa = 11.27VSNVAIASGKK145 pKa = 9.83DD146 pKa = 3.08PKK148 pKa = 11.13DD149 pKa = 3.48RR150 pKa = 11.84DD151 pKa = 3.98VIDD154 pKa = 4.22EE155 pKa = 4.36SVSGNVPEE163 pKa = 5.49AGDD166 pKa = 5.12PIDD169 pKa = 5.08PDD171 pKa = 3.84CTNCTIAPLPWKK183 pKa = 10.42VIEE186 pKa = 4.87ANNDD190 pKa = 2.67TPAPVNGKK198 pKa = 9.81DD199 pKa = 3.42GSTTPSVLDD208 pKa = 3.34NDD210 pKa = 4.49MLNGSPVIPAEE221 pKa = 4.22VTLTPGTSPHH231 pKa = 6.29TGIVMNPDD239 pKa = 2.82GTVTVSAEE247 pKa = 4.39TPAGTYY253 pKa = 7.51TYY255 pKa = 9.55PYY257 pKa = 8.94TICEE261 pKa = 4.03VLNPSNCDD269 pKa = 3.28NAVVTILVEE278 pKa = 3.93AAVIEE283 pKa = 4.5ANNDD287 pKa = 2.73TPAPVNGKK295 pKa = 9.95DD296 pKa = 3.28GGTTPSVLDD305 pKa = 3.4NDD307 pKa = 4.49MLNGSPVIPAEE318 pKa = 4.22VTLTPGTSPHH328 pKa = 6.29TGIVMNPDD336 pKa = 2.82GTVTVSAEE344 pKa = 4.39TPAGTYY350 pKa = 7.51TYY352 pKa = 9.55PYY354 pKa = 8.94TICEE358 pKa = 4.03VLNPSNCDD366 pKa = 3.28NAVVTILVEE375 pKa = 3.93AAVIEE380 pKa = 4.5ANNDD384 pKa = 2.73TPAPVNGKK392 pKa = 9.95DD393 pKa = 3.28GGTTPSVLDD402 pKa = 3.4NDD404 pKa = 4.49MLNGSPVIPSEE415 pKa = 4.21VTLTPGTSPHH425 pKa = 6.36TGITMNPDD433 pKa = 2.74GTVTVSAEE441 pKa = 4.39TPAGTYY447 pKa = 7.51TYY449 pKa = 9.55PYY451 pKa = 8.94TICEE455 pKa = 4.03VLNPSNCDD463 pKa = 3.28NAVMTIVVEE472 pKa = 4.09AAVIEE477 pKa = 4.7ANDD480 pKa = 3.96DD481 pKa = 3.42TPAPINGKK489 pKa = 10.01DD490 pKa = 3.34GGTTPSVLDD499 pKa = 3.4NDD501 pKa = 4.49MLNGSPVIPSEE512 pKa = 4.31VTLTWSPDD520 pKa = 3.42NVEE523 pKa = 4.29SPAIILNTDD532 pKa = 3.17GTVTVLPEE540 pKa = 4.18TPEE543 pKa = 4.48GIYY546 pKa = 10.16EE547 pKa = 3.87FSYY550 pKa = 10.51RR551 pKa = 11.84ICEE554 pKa = 4.03VLNPTNCDD562 pKa = 2.9EE563 pKa = 4.22AAITIVIEE571 pKa = 3.93PAPIVAMDD579 pKa = 4.66DD580 pKa = 3.79NIQIEE585 pKa = 4.26WDD587 pKa = 3.43RR588 pKa = 11.84TSIITMSVLDD598 pKa = 3.76NDD600 pKa = 4.12AFNGGSIDD608 pKa = 3.95LNEE611 pKa = 4.16VTLTPGVPSDD621 pKa = 4.58PNLSMNADD629 pKa = 3.44GTINIPASTRR639 pKa = 11.84PGTYY643 pKa = 8.8TYY645 pKa = 9.94PYY647 pKa = 8.92TICEE651 pKa = 4.03VLNPSNCSTAVATIVIEE668 pKa = 4.07ASNLFIPNVITPNGDD683 pKa = 3.27GQNDD687 pKa = 3.35RR688 pKa = 11.84FEE690 pKa = 6.18IIGSEE695 pKa = 3.82NYY697 pKa = 10.29DD698 pKa = 3.28RR699 pKa = 11.84VEE701 pKa = 3.88VTIVNRR707 pKa = 11.84WGNEE711 pKa = 2.88IFRR714 pKa = 11.84NDD716 pKa = 3.42NYY718 pKa = 11.29DD719 pKa = 3.68NNWTGRR725 pKa = 11.84GLNEE729 pKa = 3.37GTYY732 pKa = 10.27YY733 pKa = 10.89YY734 pKa = 10.06IIKK737 pKa = 9.95LYY739 pKa = 10.35KK740 pKa = 8.96GSKK743 pKa = 9.16VEE745 pKa = 3.97QHH747 pKa = 6.41KK748 pKa = 10.69GWVLIKK754 pKa = 9.01THH756 pKa = 5.97
AA1 pKa = 7.03GAVYY5 pKa = 10.26NVAVAKK11 pKa = 10.87GKK13 pKa = 10.1DD14 pKa = 3.32PKK16 pKa = 11.17DD17 pKa = 3.78RR18 pKa = 11.84DD19 pKa = 3.87VEE21 pKa = 4.51GSSTDD26 pKa = 3.66NNPLDD31 pKa = 4.25PTDD34 pKa = 4.47PDD36 pKa = 4.03NPPVDD41 pKa = 4.38PNCPEE46 pKa = 4.03CTIVEE51 pKa = 4.16IDD53 pKa = 3.38QEE55 pKa = 4.45GAIEE59 pKa = 4.32VIKK62 pKa = 9.57TADD65 pKa = 2.9LSQRR69 pKa = 11.84FRR71 pKa = 11.84YY72 pKa = 10.04AGEE75 pKa = 3.85QIAYY79 pKa = 8.48EE80 pKa = 3.96ILIKK84 pKa = 10.95NVGNVTLHH92 pKa = 6.94DD93 pKa = 4.0IKK95 pKa = 11.07VTDD98 pKa = 4.36ANADD102 pKa = 3.66DD103 pKa = 4.41VNVGNIANLAVGEE116 pKa = 4.46SVTLQAYY123 pKa = 7.37HH124 pKa = 7.49TITQADD130 pKa = 3.86MEE132 pKa = 4.55RR133 pKa = 11.84GYY135 pKa = 11.27VSNVAIASGKK145 pKa = 9.83DD146 pKa = 3.08PKK148 pKa = 11.13DD149 pKa = 3.48RR150 pKa = 11.84DD151 pKa = 3.98VIDD154 pKa = 4.22EE155 pKa = 4.36SVSGNVPEE163 pKa = 5.49AGDD166 pKa = 5.12PIDD169 pKa = 5.08PDD171 pKa = 3.84CTNCTIAPLPWKK183 pKa = 10.42VIEE186 pKa = 4.87ANNDD190 pKa = 2.67TPAPVNGKK198 pKa = 9.81DD199 pKa = 3.42GSTTPSVLDD208 pKa = 3.34NDD210 pKa = 4.49MLNGSPVIPAEE221 pKa = 4.22VTLTPGTSPHH231 pKa = 6.29TGIVMNPDD239 pKa = 2.82GTVTVSAEE247 pKa = 4.39TPAGTYY253 pKa = 7.51TYY255 pKa = 9.55PYY257 pKa = 8.94TICEE261 pKa = 4.03VLNPSNCDD269 pKa = 3.28NAVVTILVEE278 pKa = 3.93AAVIEE283 pKa = 4.5ANNDD287 pKa = 2.73TPAPVNGKK295 pKa = 9.95DD296 pKa = 3.28GGTTPSVLDD305 pKa = 3.4NDD307 pKa = 4.49MLNGSPVIPAEE318 pKa = 4.22VTLTPGTSPHH328 pKa = 6.29TGIVMNPDD336 pKa = 2.82GTVTVSAEE344 pKa = 4.39TPAGTYY350 pKa = 7.51TYY352 pKa = 9.55PYY354 pKa = 8.94TICEE358 pKa = 4.03VLNPSNCDD366 pKa = 3.28NAVVTILVEE375 pKa = 3.93AAVIEE380 pKa = 4.5ANNDD384 pKa = 2.73TPAPVNGKK392 pKa = 9.95DD393 pKa = 3.28GGTTPSVLDD402 pKa = 3.4NDD404 pKa = 4.49MLNGSPVIPSEE415 pKa = 4.21VTLTPGTSPHH425 pKa = 6.36TGITMNPDD433 pKa = 2.74GTVTVSAEE441 pKa = 4.39TPAGTYY447 pKa = 7.51TYY449 pKa = 9.55PYY451 pKa = 8.94TICEE455 pKa = 4.03VLNPSNCDD463 pKa = 3.28NAVMTIVVEE472 pKa = 4.09AAVIEE477 pKa = 4.7ANDD480 pKa = 3.96DD481 pKa = 3.42TPAPINGKK489 pKa = 10.01DD490 pKa = 3.34GGTTPSVLDD499 pKa = 3.4NDD501 pKa = 4.49MLNGSPVIPSEE512 pKa = 4.31VTLTWSPDD520 pKa = 3.42NVEE523 pKa = 4.29SPAIILNTDD532 pKa = 3.17GTVTVLPEE540 pKa = 4.18TPEE543 pKa = 4.48GIYY546 pKa = 10.16EE547 pKa = 3.87FSYY550 pKa = 10.51RR551 pKa = 11.84ICEE554 pKa = 4.03VLNPTNCDD562 pKa = 2.9EE563 pKa = 4.22AAITIVIEE571 pKa = 3.93PAPIVAMDD579 pKa = 4.66DD580 pKa = 3.79NIQIEE585 pKa = 4.26WDD587 pKa = 3.43RR588 pKa = 11.84TSIITMSVLDD598 pKa = 3.76NDD600 pKa = 4.12AFNGGSIDD608 pKa = 3.95LNEE611 pKa = 4.16VTLTPGVPSDD621 pKa = 4.58PNLSMNADD629 pKa = 3.44GTINIPASTRR639 pKa = 11.84PGTYY643 pKa = 8.8TYY645 pKa = 9.94PYY647 pKa = 8.92TICEE651 pKa = 4.03VLNPSNCSTAVATIVIEE668 pKa = 4.07ASNLFIPNVITPNGDD683 pKa = 3.27GQNDD687 pKa = 3.35RR688 pKa = 11.84FEE690 pKa = 6.18IIGSEE695 pKa = 3.82NYY697 pKa = 10.29DD698 pKa = 3.28RR699 pKa = 11.84VEE701 pKa = 3.88VTIVNRR707 pKa = 11.84WGNEE711 pKa = 2.88IFRR714 pKa = 11.84NDD716 pKa = 3.42NYY718 pKa = 11.29DD719 pKa = 3.68NNWTGRR725 pKa = 11.84GLNEE729 pKa = 3.37GTYY732 pKa = 10.27YY733 pKa = 10.89YY734 pKa = 10.06IIKK737 pKa = 9.95LYY739 pKa = 10.35KK740 pKa = 8.96GSKK743 pKa = 9.16VEE745 pKa = 3.97QHH747 pKa = 6.41KK748 pKa = 10.69GWVLIKK754 pKa = 9.01THH756 pKa = 5.97
Molecular weight: 79.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U0P0R3|A0A4U0P0R3_9SPHI GtrA family protein OS=Sphingobacterium olei OX=2571155 GN=FAZ15_12560 PE=3 SV=1
MM1 pKa = 7.5SGSIHH6 pKa = 6.15PTRR9 pKa = 11.84DD10 pKa = 3.03DD11 pKa = 5.03AYY13 pKa = 10.44FKK15 pKa = 11.08PNDD18 pKa = 3.53IPHH21 pKa = 6.68LKK23 pKa = 10.38EE24 pKa = 4.58SICHH28 pKa = 5.5NPAGISPEE36 pKa = 4.02RR37 pKa = 11.84NEE39 pKa = 3.72VRR41 pKa = 11.84PNTGSVLLNRR51 pKa = 11.84SDD53 pKa = 3.89APPIPRR59 pKa = 11.84SIHH62 pKa = 5.38FLRR65 pKa = 11.84NDD67 pKa = 3.13VPLKK71 pKa = 10.26RR72 pKa = 11.84NEE74 pKa = 3.96VRR76 pKa = 11.84PKK78 pKa = 10.5ARR80 pKa = 11.84NIFLNTPSVRR90 pKa = 11.84HH91 pKa = 5.55MPQSIYY97 pKa = 9.7FVRR100 pKa = 11.84VNVHH104 pKa = 6.12HH105 pKa = 6.6KK106 pKa = 10.93RR107 pKa = 11.84NDD109 pKa = 3.16GSLNAGRR116 pKa = 11.84VFLNPPSVCHH126 pKa = 6.43IPQSIYY132 pKa = 9.36FVRR135 pKa = 11.84VNVYY139 pKa = 10.2HH140 pKa = 7.24KK141 pKa = 10.99
MM1 pKa = 7.5SGSIHH6 pKa = 6.15PTRR9 pKa = 11.84DD10 pKa = 3.03DD11 pKa = 5.03AYY13 pKa = 10.44FKK15 pKa = 11.08PNDD18 pKa = 3.53IPHH21 pKa = 6.68LKK23 pKa = 10.38EE24 pKa = 4.58SICHH28 pKa = 5.5NPAGISPEE36 pKa = 4.02RR37 pKa = 11.84NEE39 pKa = 3.72VRR41 pKa = 11.84PNTGSVLLNRR51 pKa = 11.84SDD53 pKa = 3.89APPIPRR59 pKa = 11.84SIHH62 pKa = 5.38FLRR65 pKa = 11.84NDD67 pKa = 3.13VPLKK71 pKa = 10.26RR72 pKa = 11.84NEE74 pKa = 3.96VRR76 pKa = 11.84PKK78 pKa = 10.5ARR80 pKa = 11.84NIFLNTPSVRR90 pKa = 11.84HH91 pKa = 5.55MPQSIYY97 pKa = 9.7FVRR100 pKa = 11.84VNVHH104 pKa = 6.12HH105 pKa = 6.6KK106 pKa = 10.93RR107 pKa = 11.84NDD109 pKa = 3.16GSLNAGRR116 pKa = 11.84VFLNPPSVCHH126 pKa = 6.43IPQSIYY132 pKa = 9.36FVRR135 pKa = 11.84VNVYY139 pKa = 10.2HH140 pKa = 7.24KK141 pKa = 10.99
Molecular weight: 16.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1597812 |
20 |
3026 |
368.1 |
41.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.965 ± 0.032 | 0.695 ± 0.01 |
5.638 ± 0.022 | 6.052 ± 0.032 |
4.838 ± 0.025 | 6.859 ± 0.035 |
1.944 ± 0.018 | 7.306 ± 0.033 |
6.425 ± 0.039 | 9.398 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.197 ± 0.013 | 5.673 ± 0.033 |
3.656 ± 0.021 | 3.764 ± 0.023 |
4.275 ± 0.026 | 6.562 ± 0.024 |
5.722 ± 0.032 | 6.401 ± 0.027 |
1.26 ± 0.015 | 4.371 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
