
Shrew coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; unclassified Orthocoronavirinae
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
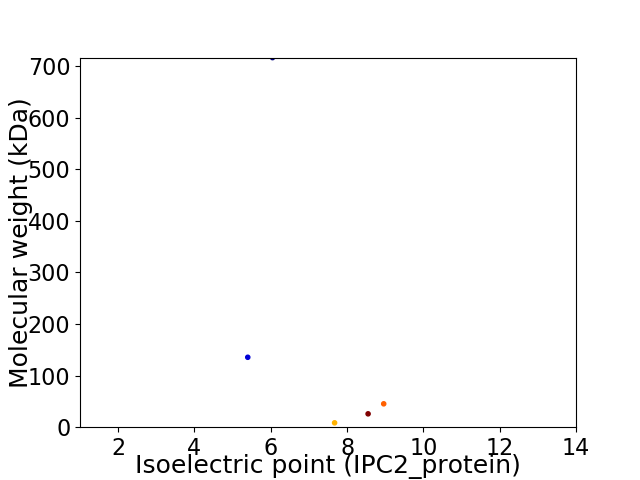
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4MZ81|A0A2H4MZ81_9NIDO 2'-O-methyltransferase OS=Shrew coronavirus OX=2050019 PE=3 SV=1
MM1 pKa = 7.76LILVFLSLVTVSLQNVVVPFDD22 pKa = 3.68NYY24 pKa = 10.96CNLDD28 pKa = 3.34ARR30 pKa = 11.84YY31 pKa = 10.07HH32 pKa = 6.2NITVQSEE39 pKa = 4.28KK40 pKa = 10.58PFNFNATFYY49 pKa = 7.63TTNQTCFDD57 pKa = 4.17VNCTSSNTSNTFNSSDD73 pKa = 3.55TITYY77 pKa = 9.29SNCSSVTFNGCVVKK91 pKa = 9.04QFCVTQPCHH100 pKa = 6.92CYY102 pKa = 10.34YY103 pKa = 9.58QTQQVSGFVFDD114 pKa = 4.53GLSHH118 pKa = 6.28TFVPEE123 pKa = 3.53ISNYY127 pKa = 9.62HH128 pKa = 5.48VADD131 pKa = 3.85VVNKK135 pKa = 9.29TLTRR139 pKa = 11.84TTNLCSSVNVTIDD152 pKa = 3.29YY153 pKa = 10.49KK154 pKa = 10.83QFLMLSSVEE163 pKa = 4.06INSTFVVTQFVPSYY177 pKa = 11.11VYY179 pKa = 10.81NQDD182 pKa = 2.79NFTIKK187 pKa = 10.63GDD189 pKa = 3.96NFWVVANTTTVFVLQPKK206 pKa = 8.62TPSNEE211 pKa = 4.08TLSLYY216 pKa = 10.32IKK218 pKa = 9.08ITNSSGSLFVNVTICNYY235 pKa = 9.65FRR237 pKa = 11.84VLNNTDD243 pKa = 3.21NTNTSSLVYY252 pKa = 8.51TNCVFEE258 pKa = 4.47KK259 pKa = 10.71VAGGLQVVTLGVIWRR274 pKa = 11.84KK275 pKa = 9.55NVVTLHH281 pKa = 6.04WLAQEE286 pKa = 4.29VVTIEE291 pKa = 5.42LPFDD295 pKa = 3.92PQWYY299 pKa = 10.12GGDD302 pKa = 3.46PALNGLYY309 pKa = 10.72CNGWYY314 pKa = 10.42ANSLEE319 pKa = 4.32SPHH322 pKa = 6.77TYY324 pKa = 10.59LLKK327 pKa = 10.23TNASGHH333 pKa = 5.7IVNSLWCNPDD343 pKa = 4.07DD344 pKa = 4.33EE345 pKa = 5.72LSSLQCAYY353 pKa = 9.07KK354 pKa = 10.25TFNLNRR360 pKa = 11.84GVYY363 pKa = 10.01SRR365 pKa = 11.84TVHH368 pKa = 5.56QKK370 pKa = 9.39VVYY373 pKa = 9.23LQRR376 pKa = 11.84FSSTPKK382 pKa = 10.37CVTNKK387 pKa = 9.55VNADD391 pKa = 3.49LVVRR395 pKa = 11.84GGFYY399 pKa = 10.07ATLTNCDD406 pKa = 3.52YY407 pKa = 11.37DD408 pKa = 3.82LHH410 pKa = 7.26GLLNISLLPQRR421 pKa = 11.84WGGVNYY427 pKa = 7.78TIPPVVVPWVLKK439 pKa = 11.06NPDD442 pKa = 3.1GTSRR446 pKa = 11.84LWQQCVGVDD455 pKa = 3.39VASANTQCFKK465 pKa = 10.82SISVSFVTNAYY476 pKa = 10.37LLEE479 pKa = 4.55LLPGQCVSYY488 pKa = 10.72YY489 pKa = 10.98GYY491 pKa = 8.63FTTGAAGLVAKK502 pKa = 10.24NAIVGTFDD510 pKa = 4.4LSSGDD515 pKa = 3.84TQTCMYY521 pKa = 9.23QQQNTTFEE529 pKa = 4.27IPQNVCVSFTFDD541 pKa = 2.91SQTIKK546 pKa = 11.22GILTQFNTTYY556 pKa = 10.2PSNYY560 pKa = 9.31TNILKK565 pKa = 10.7HH566 pKa = 6.0NGVVKK571 pKa = 9.11YY572 pKa = 10.53VRR574 pKa = 11.84IDD576 pKa = 3.32GVSYY580 pKa = 10.29VVKK583 pKa = 9.69PCLFEE588 pKa = 4.49SVVKK592 pKa = 10.45IVNNATYY599 pKa = 10.09NHH601 pKa = 6.96TIISTSLCGEE611 pKa = 4.05NGTNTTIGCIQTGFVNEE628 pKa = 4.23TLNTCDD634 pKa = 3.49YY635 pKa = 11.18SLGNGFCANVNKK647 pKa = 10.8SMDD650 pKa = 3.5VYY652 pKa = 11.34KK653 pKa = 10.42FIPSKK658 pKa = 9.21QTNDD662 pKa = 3.26YY663 pKa = 9.71VVPISGMYY671 pKa = 10.53NITLPTSYY679 pKa = 11.14EE680 pKa = 4.12PFTSTAYY687 pKa = 10.14YY688 pKa = 7.27QTRR691 pKa = 11.84LEE693 pKa = 4.5KK694 pKa = 10.89LQVDD698 pKa = 4.45CQHH701 pKa = 6.4FVCDD705 pKa = 4.58SNIHH709 pKa = 6.43CLTLLQKK716 pKa = 11.12YY717 pKa = 10.68GDD719 pKa = 3.92ICTKK723 pKa = 9.28ITGDD727 pKa = 3.72VNTVMASLTSLEE739 pKa = 4.29LDD741 pKa = 3.42ALQTVALEE749 pKa = 4.21GLNVDD754 pKa = 3.48YY755 pKa = 11.4GSFNFSGLMPQRR767 pKa = 11.84GTRR770 pKa = 11.84SFIEE774 pKa = 4.61DD775 pKa = 3.87LLFDD779 pKa = 5.23KK780 pKa = 10.73IVTTGPQFYY789 pKa = 10.3KK790 pKa = 10.46DD791 pKa = 4.14YY792 pKa = 10.92YY793 pKa = 10.63DD794 pKa = 3.49CHH796 pKa = 7.18KK797 pKa = 11.1DD798 pKa = 3.19KK799 pKa = 10.96TINDD803 pKa = 4.53LLCAQQYY810 pKa = 11.09NGMLVVPPIMDD821 pKa = 4.59DD822 pKa = 3.21ATIAMYY828 pKa = 8.41GTIAGASVSAGMFGGQAGMITWNTAVSARR857 pKa = 11.84LNALGITQEE866 pKa = 4.17VMQSDD871 pKa = 3.53IQKK874 pKa = 9.95IANAVNGIVDD884 pKa = 3.88HH885 pKa = 6.39VSNLAKK891 pKa = 9.74TVAGGFEE898 pKa = 4.4VIQAVVNQNGAQMMALVDD916 pKa = 3.81GLQEE920 pKa = 3.97NFGAISNNFEE930 pKa = 4.45TINNRR935 pKa = 11.84LNKK938 pKa = 10.35LEE940 pKa = 5.38ADD942 pKa = 3.51QQMDD946 pKa = 3.53RR947 pKa = 11.84LINGRR952 pKa = 11.84LNVLQSFMTNYY963 pKa = 9.61KK964 pKa = 9.75IKK966 pKa = 10.37VSAMQATQHH975 pKa = 5.35TVNSIIKK982 pKa = 8.35EE983 pKa = 4.06CVYY986 pKa = 10.88AQSTRR991 pKa = 11.84NGFCGDD997 pKa = 3.67GKK999 pKa = 10.21HH1000 pKa = 6.37IMSFMQNAPDD1010 pKa = 4.7GIFFIHH1016 pKa = 6.23FTMKK1020 pKa = 9.43PVQYY1024 pKa = 10.42INVTTTPGLCINNTGIAPKK1043 pKa = 10.42DD1044 pKa = 3.69GIFVFVNGTWMVSPRR1059 pKa = 11.84NIYY1062 pKa = 10.18NPRR1065 pKa = 11.84NVSNSDD1071 pKa = 2.62IVHH1074 pKa = 6.34YY1075 pKa = 10.22ISCEE1079 pKa = 3.73ANYY1082 pKa = 9.76TLVNNTIGSVVLPQLPNIGDD1102 pKa = 3.93DD1103 pKa = 4.1FNNLYY1108 pKa = 10.89EE1109 pKa = 4.31NTSKK1113 pKa = 11.11ALEE1116 pKa = 4.46EE1117 pKa = 3.92IKK1119 pKa = 10.76NIYY1122 pKa = 10.52FNFTNLNLTTEE1133 pKa = 4.37IEE1135 pKa = 4.24RR1136 pKa = 11.84LNSIAEE1142 pKa = 4.19EE1143 pKa = 4.2VKK1145 pKa = 10.64KK1146 pKa = 11.19LNITLSEE1153 pKa = 4.03LGRR1156 pKa = 11.84YY1157 pKa = 7.15EE1158 pKa = 6.13RR1159 pKa = 11.84YY1160 pKa = 10.21VKK1162 pKa = 9.79WPWYY1166 pKa = 8.27VWLAIAVVMIVLVFILGCLLFMTGCCGCCGCCGGCCGKK1204 pKa = 10.47ACSKK1208 pKa = 10.62PPDD1211 pKa = 3.64YY1212 pKa = 11.43EE1213 pKa = 4.09EE1214 pKa = 5.02LEE1216 pKa = 4.36KK1217 pKa = 11.27LHH1219 pKa = 6.35VAA1221 pKa = 4.38
MM1 pKa = 7.76LILVFLSLVTVSLQNVVVPFDD22 pKa = 3.68NYY24 pKa = 10.96CNLDD28 pKa = 3.34ARR30 pKa = 11.84YY31 pKa = 10.07HH32 pKa = 6.2NITVQSEE39 pKa = 4.28KK40 pKa = 10.58PFNFNATFYY49 pKa = 7.63TTNQTCFDD57 pKa = 4.17VNCTSSNTSNTFNSSDD73 pKa = 3.55TITYY77 pKa = 9.29SNCSSVTFNGCVVKK91 pKa = 9.04QFCVTQPCHH100 pKa = 6.92CYY102 pKa = 10.34YY103 pKa = 9.58QTQQVSGFVFDD114 pKa = 4.53GLSHH118 pKa = 6.28TFVPEE123 pKa = 3.53ISNYY127 pKa = 9.62HH128 pKa = 5.48VADD131 pKa = 3.85VVNKK135 pKa = 9.29TLTRR139 pKa = 11.84TTNLCSSVNVTIDD152 pKa = 3.29YY153 pKa = 10.49KK154 pKa = 10.83QFLMLSSVEE163 pKa = 4.06INSTFVVTQFVPSYY177 pKa = 11.11VYY179 pKa = 10.81NQDD182 pKa = 2.79NFTIKK187 pKa = 10.63GDD189 pKa = 3.96NFWVVANTTTVFVLQPKK206 pKa = 8.62TPSNEE211 pKa = 4.08TLSLYY216 pKa = 10.32IKK218 pKa = 9.08ITNSSGSLFVNVTICNYY235 pKa = 9.65FRR237 pKa = 11.84VLNNTDD243 pKa = 3.21NTNTSSLVYY252 pKa = 8.51TNCVFEE258 pKa = 4.47KK259 pKa = 10.71VAGGLQVVTLGVIWRR274 pKa = 11.84KK275 pKa = 9.55NVVTLHH281 pKa = 6.04WLAQEE286 pKa = 4.29VVTIEE291 pKa = 5.42LPFDD295 pKa = 3.92PQWYY299 pKa = 10.12GGDD302 pKa = 3.46PALNGLYY309 pKa = 10.72CNGWYY314 pKa = 10.42ANSLEE319 pKa = 4.32SPHH322 pKa = 6.77TYY324 pKa = 10.59LLKK327 pKa = 10.23TNASGHH333 pKa = 5.7IVNSLWCNPDD343 pKa = 4.07DD344 pKa = 4.33EE345 pKa = 5.72LSSLQCAYY353 pKa = 9.07KK354 pKa = 10.25TFNLNRR360 pKa = 11.84GVYY363 pKa = 10.01SRR365 pKa = 11.84TVHH368 pKa = 5.56QKK370 pKa = 9.39VVYY373 pKa = 9.23LQRR376 pKa = 11.84FSSTPKK382 pKa = 10.37CVTNKK387 pKa = 9.55VNADD391 pKa = 3.49LVVRR395 pKa = 11.84GGFYY399 pKa = 10.07ATLTNCDD406 pKa = 3.52YY407 pKa = 11.37DD408 pKa = 3.82LHH410 pKa = 7.26GLLNISLLPQRR421 pKa = 11.84WGGVNYY427 pKa = 7.78TIPPVVVPWVLKK439 pKa = 11.06NPDD442 pKa = 3.1GTSRR446 pKa = 11.84LWQQCVGVDD455 pKa = 3.39VASANTQCFKK465 pKa = 10.82SISVSFVTNAYY476 pKa = 10.37LLEE479 pKa = 4.55LLPGQCVSYY488 pKa = 10.72YY489 pKa = 10.98GYY491 pKa = 8.63FTTGAAGLVAKK502 pKa = 10.24NAIVGTFDD510 pKa = 4.4LSSGDD515 pKa = 3.84TQTCMYY521 pKa = 9.23QQQNTTFEE529 pKa = 4.27IPQNVCVSFTFDD541 pKa = 2.91SQTIKK546 pKa = 11.22GILTQFNTTYY556 pKa = 10.2PSNYY560 pKa = 9.31TNILKK565 pKa = 10.7HH566 pKa = 6.0NGVVKK571 pKa = 9.11YY572 pKa = 10.53VRR574 pKa = 11.84IDD576 pKa = 3.32GVSYY580 pKa = 10.29VVKK583 pKa = 9.69PCLFEE588 pKa = 4.49SVVKK592 pKa = 10.45IVNNATYY599 pKa = 10.09NHH601 pKa = 6.96TIISTSLCGEE611 pKa = 4.05NGTNTTIGCIQTGFVNEE628 pKa = 4.23TLNTCDD634 pKa = 3.49YY635 pKa = 11.18SLGNGFCANVNKK647 pKa = 10.8SMDD650 pKa = 3.5VYY652 pKa = 11.34KK653 pKa = 10.42FIPSKK658 pKa = 9.21QTNDD662 pKa = 3.26YY663 pKa = 9.71VVPISGMYY671 pKa = 10.53NITLPTSYY679 pKa = 11.14EE680 pKa = 4.12PFTSTAYY687 pKa = 10.14YY688 pKa = 7.27QTRR691 pKa = 11.84LEE693 pKa = 4.5KK694 pKa = 10.89LQVDD698 pKa = 4.45CQHH701 pKa = 6.4FVCDD705 pKa = 4.58SNIHH709 pKa = 6.43CLTLLQKK716 pKa = 11.12YY717 pKa = 10.68GDD719 pKa = 3.92ICTKK723 pKa = 9.28ITGDD727 pKa = 3.72VNTVMASLTSLEE739 pKa = 4.29LDD741 pKa = 3.42ALQTVALEE749 pKa = 4.21GLNVDD754 pKa = 3.48YY755 pKa = 11.4GSFNFSGLMPQRR767 pKa = 11.84GTRR770 pKa = 11.84SFIEE774 pKa = 4.61DD775 pKa = 3.87LLFDD779 pKa = 5.23KK780 pKa = 10.73IVTTGPQFYY789 pKa = 10.3KK790 pKa = 10.46DD791 pKa = 4.14YY792 pKa = 10.92YY793 pKa = 10.63DD794 pKa = 3.49CHH796 pKa = 7.18KK797 pKa = 11.1DD798 pKa = 3.19KK799 pKa = 10.96TINDD803 pKa = 4.53LLCAQQYY810 pKa = 11.09NGMLVVPPIMDD821 pKa = 4.59DD822 pKa = 3.21ATIAMYY828 pKa = 8.41GTIAGASVSAGMFGGQAGMITWNTAVSARR857 pKa = 11.84LNALGITQEE866 pKa = 4.17VMQSDD871 pKa = 3.53IQKK874 pKa = 9.95IANAVNGIVDD884 pKa = 3.88HH885 pKa = 6.39VSNLAKK891 pKa = 9.74TVAGGFEE898 pKa = 4.4VIQAVVNQNGAQMMALVDD916 pKa = 3.81GLQEE920 pKa = 3.97NFGAISNNFEE930 pKa = 4.45TINNRR935 pKa = 11.84LNKK938 pKa = 10.35LEE940 pKa = 5.38ADD942 pKa = 3.51QQMDD946 pKa = 3.53RR947 pKa = 11.84LINGRR952 pKa = 11.84LNVLQSFMTNYY963 pKa = 9.61KK964 pKa = 9.75IKK966 pKa = 10.37VSAMQATQHH975 pKa = 5.35TVNSIIKK982 pKa = 8.35EE983 pKa = 4.06CVYY986 pKa = 10.88AQSTRR991 pKa = 11.84NGFCGDD997 pKa = 3.67GKK999 pKa = 10.21HH1000 pKa = 6.37IMSFMQNAPDD1010 pKa = 4.7GIFFIHH1016 pKa = 6.23FTMKK1020 pKa = 9.43PVQYY1024 pKa = 10.42INVTTTPGLCINNTGIAPKK1043 pKa = 10.42DD1044 pKa = 3.69GIFVFVNGTWMVSPRR1059 pKa = 11.84NIYY1062 pKa = 10.18NPRR1065 pKa = 11.84NVSNSDD1071 pKa = 2.62IVHH1074 pKa = 6.34YY1075 pKa = 10.22ISCEE1079 pKa = 3.73ANYY1082 pKa = 9.76TLVNNTIGSVVLPQLPNIGDD1102 pKa = 3.93DD1103 pKa = 4.1FNNLYY1108 pKa = 10.89EE1109 pKa = 4.31NTSKK1113 pKa = 11.11ALEE1116 pKa = 4.46EE1117 pKa = 3.92IKK1119 pKa = 10.76NIYY1122 pKa = 10.52FNFTNLNLTTEE1133 pKa = 4.37IEE1135 pKa = 4.24RR1136 pKa = 11.84LNSIAEE1142 pKa = 4.19EE1143 pKa = 4.2VKK1145 pKa = 10.64KK1146 pKa = 11.19LNITLSEE1153 pKa = 4.03LGRR1156 pKa = 11.84YY1157 pKa = 7.15EE1158 pKa = 6.13RR1159 pKa = 11.84YY1160 pKa = 10.21VKK1162 pKa = 9.79WPWYY1166 pKa = 8.27VWLAIAVVMIVLVFILGCLLFMTGCCGCCGCCGGCCGKK1204 pKa = 10.47ACSKK1208 pKa = 10.62PPDD1211 pKa = 3.64YY1212 pKa = 11.43EE1213 pKa = 4.09EE1214 pKa = 5.02LEE1216 pKa = 4.36KK1217 pKa = 11.27LHH1219 pKa = 6.35VAA1221 pKa = 4.38
Molecular weight: 135.55 kDa
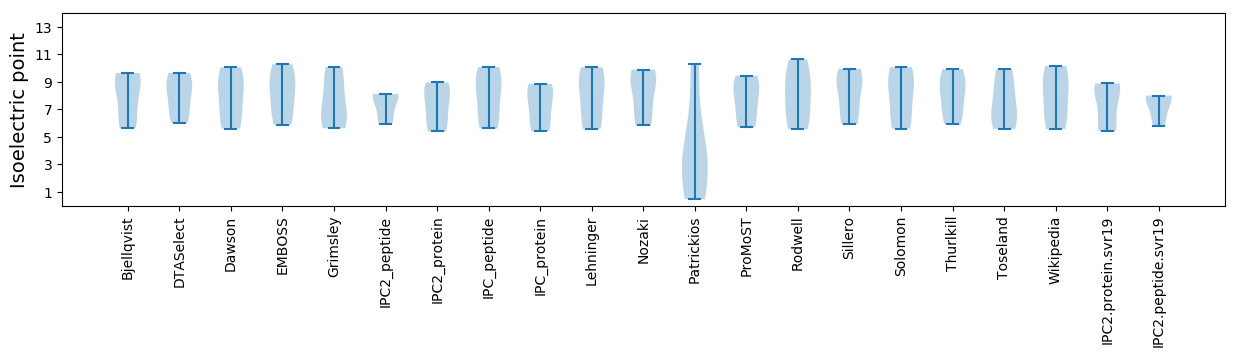
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4MXZ6|A0A2H4MXZ6_9NIDO Spike glycoprotein OS=Shrew coronavirus OX=2050019 PE=4 SV=1
MM1 pKa = 7.42SGKK4 pKa = 9.49SWADD8 pKa = 3.14RR9 pKa = 11.84VEE11 pKa = 5.07AEE13 pKa = 4.1EE14 pKa = 4.52QKK16 pKa = 11.05NKK18 pKa = 8.66PQQRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84SKK27 pKa = 10.26SKK29 pKa = 10.49DD30 pKa = 3.17RR31 pKa = 11.84KK32 pKa = 8.51TMPLSWFNPLEE43 pKa = 4.07FEE45 pKa = 4.36VGKK48 pKa = 10.5DD49 pKa = 3.38LGDD52 pKa = 4.94LINTNSVPIGKK63 pKa = 8.21GTKK66 pKa = 8.95QEE68 pKa = 3.79QHH70 pKa = 6.75GYY72 pKa = 9.68WNIQTRR78 pKa = 11.84FRR80 pKa = 11.84VNKK83 pKa = 9.47GKK85 pKa = 10.45RR86 pKa = 11.84VDD88 pKa = 3.76LQPRR92 pKa = 11.84AFFYY96 pKa = 9.94YY97 pKa = 10.17TGTGPHH103 pKa = 7.06KK104 pKa = 10.26ILAFGEE110 pKa = 4.22QQEE113 pKa = 4.58GVVWVATKK121 pKa = 10.32EE122 pKa = 3.91ADD124 pKa = 3.45HH125 pKa = 6.46NPVRR129 pKa = 11.84FGDD132 pKa = 3.91RR133 pKa = 11.84PEE135 pKa = 4.08STPAMEE141 pKa = 4.18VKK143 pKa = 10.15IRR145 pKa = 11.84QGKK148 pKa = 8.33QPLGVKK154 pKa = 9.97LLNDD158 pKa = 3.45SKK160 pKa = 11.49EE161 pKa = 4.07EE162 pKa = 3.86VVRR165 pKa = 11.84QSRR168 pKa = 11.84ARR170 pKa = 11.84SRR172 pKa = 11.84SRR174 pKa = 11.84SQSRR178 pKa = 11.84SRR180 pKa = 11.84DD181 pKa = 3.37TSQKK185 pKa = 10.22RR186 pKa = 11.84VTFSDD191 pKa = 3.54QQPKK195 pKa = 9.8GDD197 pKa = 3.55QYY199 pKa = 10.18VTKK202 pKa = 10.74NDD204 pKa = 3.61LSNLLSKK211 pKa = 11.04LLDD214 pKa = 3.61EE215 pKa = 5.35KK216 pKa = 11.33LNSNPKK222 pKa = 8.87PQRR225 pKa = 11.84QPRR228 pKa = 11.84SRR230 pKa = 11.84SNSSTRR236 pKa = 11.84NSQKK240 pKa = 8.91EE241 pKa = 4.13TQAVQKK247 pKa = 10.92DD248 pKa = 3.91QMSKK252 pKa = 9.63HH253 pKa = 3.78WWKK256 pKa = 10.76RR257 pKa = 11.84IPTKK261 pKa = 10.83DD262 pKa = 3.47EE263 pKa = 4.76GIEE266 pKa = 3.73QCFGQRR272 pKa = 11.84SDD274 pKa = 3.33TVNFGTKK281 pKa = 10.5YY282 pKa = 9.89MVDD285 pKa = 3.04QGTGGNFPQLATLLPTPAAMLYY307 pKa = 9.89GSHH310 pKa = 5.76VQITPGMSPDD320 pKa = 3.52KK321 pKa = 10.79EE322 pKa = 4.28FIVYY326 pKa = 8.65TCGIEE331 pKa = 3.95VDD333 pKa = 4.2KK334 pKa = 11.37NDD336 pKa = 5.12PIYY339 pKa = 11.05KK340 pKa = 9.61EE341 pKa = 3.92FKK343 pKa = 10.17KK344 pKa = 10.85GVNAFKK350 pKa = 10.86DD351 pKa = 3.54DD352 pKa = 4.02TTWLSGNTIAKK363 pKa = 9.8AKK365 pKa = 10.07NLNTTSTPSQTVQSVSEE382 pKa = 4.39SNASVVVKK390 pKa = 10.29FDD392 pKa = 3.77EE393 pKa = 4.51NGEE396 pKa = 4.34EE397 pKa = 4.28VSSAA401 pKa = 3.28
MM1 pKa = 7.42SGKK4 pKa = 9.49SWADD8 pKa = 3.14RR9 pKa = 11.84VEE11 pKa = 5.07AEE13 pKa = 4.1EE14 pKa = 4.52QKK16 pKa = 11.05NKK18 pKa = 8.66PQQRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84SKK27 pKa = 10.26SKK29 pKa = 10.49DD30 pKa = 3.17RR31 pKa = 11.84KK32 pKa = 8.51TMPLSWFNPLEE43 pKa = 4.07FEE45 pKa = 4.36VGKK48 pKa = 10.5DD49 pKa = 3.38LGDD52 pKa = 4.94LINTNSVPIGKK63 pKa = 8.21GTKK66 pKa = 8.95QEE68 pKa = 3.79QHH70 pKa = 6.75GYY72 pKa = 9.68WNIQTRR78 pKa = 11.84FRR80 pKa = 11.84VNKK83 pKa = 9.47GKK85 pKa = 10.45RR86 pKa = 11.84VDD88 pKa = 3.76LQPRR92 pKa = 11.84AFFYY96 pKa = 9.94YY97 pKa = 10.17TGTGPHH103 pKa = 7.06KK104 pKa = 10.26ILAFGEE110 pKa = 4.22QQEE113 pKa = 4.58GVVWVATKK121 pKa = 10.32EE122 pKa = 3.91ADD124 pKa = 3.45HH125 pKa = 6.46NPVRR129 pKa = 11.84FGDD132 pKa = 3.91RR133 pKa = 11.84PEE135 pKa = 4.08STPAMEE141 pKa = 4.18VKK143 pKa = 10.15IRR145 pKa = 11.84QGKK148 pKa = 8.33QPLGVKK154 pKa = 9.97LLNDD158 pKa = 3.45SKK160 pKa = 11.49EE161 pKa = 4.07EE162 pKa = 3.86VVRR165 pKa = 11.84QSRR168 pKa = 11.84ARR170 pKa = 11.84SRR172 pKa = 11.84SRR174 pKa = 11.84SQSRR178 pKa = 11.84SRR180 pKa = 11.84DD181 pKa = 3.37TSQKK185 pKa = 10.22RR186 pKa = 11.84VTFSDD191 pKa = 3.54QQPKK195 pKa = 9.8GDD197 pKa = 3.55QYY199 pKa = 10.18VTKK202 pKa = 10.74NDD204 pKa = 3.61LSNLLSKK211 pKa = 11.04LLDD214 pKa = 3.61EE215 pKa = 5.35KK216 pKa = 11.33LNSNPKK222 pKa = 8.87PQRR225 pKa = 11.84QPRR228 pKa = 11.84SRR230 pKa = 11.84SNSSTRR236 pKa = 11.84NSQKK240 pKa = 8.91EE241 pKa = 4.13TQAVQKK247 pKa = 10.92DD248 pKa = 3.91QMSKK252 pKa = 9.63HH253 pKa = 3.78WWKK256 pKa = 10.76RR257 pKa = 11.84IPTKK261 pKa = 10.83DD262 pKa = 3.47EE263 pKa = 4.76GIEE266 pKa = 3.73QCFGQRR272 pKa = 11.84SDD274 pKa = 3.33TVNFGTKK281 pKa = 10.5YY282 pKa = 9.89MVDD285 pKa = 3.04QGTGGNFPQLATLLPTPAAMLYY307 pKa = 9.89GSHH310 pKa = 5.76VQITPGMSPDD320 pKa = 3.52KK321 pKa = 10.79EE322 pKa = 4.28FIVYY326 pKa = 8.65TCGIEE331 pKa = 3.95VDD333 pKa = 4.2KK334 pKa = 11.37NDD336 pKa = 5.12PIYY339 pKa = 11.05KK340 pKa = 9.61EE341 pKa = 3.92FKK343 pKa = 10.17KK344 pKa = 10.85GVNAFKK350 pKa = 10.86DD351 pKa = 3.54DD352 pKa = 4.02TTWLSGNTIAKK363 pKa = 9.8AKK365 pKa = 10.07NLNTTSTPSQTVQSVSEE382 pKa = 4.39SNASVVVKK390 pKa = 10.29FDD392 pKa = 3.77EE393 pKa = 4.51NGEE396 pKa = 4.34EE397 pKa = 4.28VSSAA401 pKa = 3.28
Molecular weight: 45.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8310 |
78 |
6381 |
1662.0 |
186.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.199 ± 0.358 | 3.634 ± 0.694 |
5.884 ± 0.656 | 3.911 ± 0.594 |
5.199 ± 0.441 | 5.716 ± 0.323 |
1.757 ± 0.131 | 4.862 ± 1.332 |
6.751 ± 0.966 | 8.448 ± 0.67 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.118 ± 0.14 | 6.258 ± 1.003 |
3.682 ± 0.299 | 3.502 ± 0.868 |
2.78 ± 0.703 | 6.955 ± 0.55 |
7.064 ± 0.668 | 10.301 ± 0.598 |
1.372 ± 0.238 | 4.609 ± 0.532 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
