
Aspergillus ellipticus CBS 707.79
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus ellipticus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
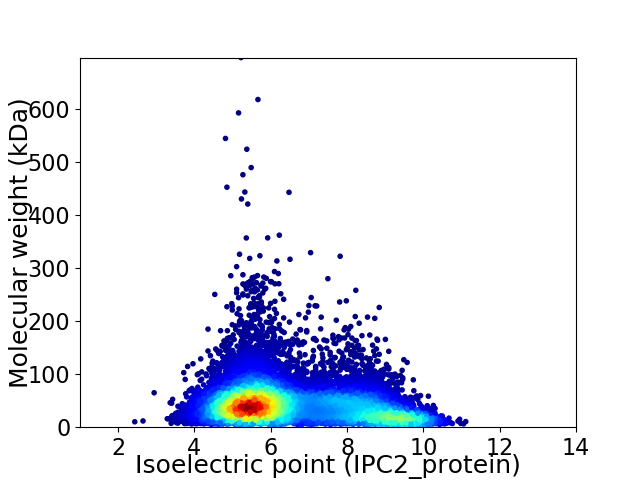
Virtual 2D-PAGE plot for 12861 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A319D1A5|A0A319D1A5_9EURO Uncharacterized protein OS=Aspergillus ellipticus CBS 707.79 OX=1448320 GN=BO71DRAFT_67798 PE=4 SV=1
MM1 pKa = 8.03GDD3 pKa = 3.49TFHH6 pKa = 7.89PDD8 pKa = 3.22NSWSLLDD15 pKa = 5.4DD16 pKa = 4.08DD17 pKa = 4.71TPAYY21 pKa = 10.43DD22 pKa = 5.15DD23 pKa = 3.99YY24 pKa = 11.88PEE26 pKa = 4.73VIPLATTEE34 pKa = 4.17YY35 pKa = 10.63VDD37 pKa = 4.1PFPAPFSTQGYY48 pKa = 8.69APRR51 pKa = 11.84EE52 pKa = 4.0IIQAVWSVSGAPSNQNIPAAAPVPNTNPDD81 pKa = 3.37PAAAPVIDD89 pKa = 4.48PNLNQTAAPTVDD101 pKa = 3.59SNLNNTEE108 pKa = 4.02APVVDD113 pKa = 4.04PGLYY117 pKa = 9.92PSVAPGTTFYY127 pKa = 10.79FDD129 pKa = 3.23QVVGVFTGSDD139 pKa = 3.98LDD141 pKa = 3.56PTMPLFLEE149 pKa = 4.88PDD151 pKa = 2.98WSFPPLIQSMPPGVSPTADD170 pKa = 3.45ATPTAAVDD178 pKa = 3.51ATMDD182 pKa = 4.0PAIDD186 pKa = 4.1PSQDD190 pKa = 3.03VAADD194 pKa = 3.67PAIDD198 pKa = 4.11PSQDD202 pKa = 3.03VAADD206 pKa = 3.64PAIDD210 pKa = 4.26PNLDD214 pKa = 3.03AAADD218 pKa = 4.0PNIDD222 pKa = 3.91PNLDD226 pKa = 3.12AAADD230 pKa = 4.08PNIHH234 pKa = 6.89SDD236 pKa = 3.99LDD238 pKa = 4.16LAALWAEE245 pKa = 4.67LDD247 pKa = 3.43QTAAPGNDD255 pKa = 3.05TGPAQTVVPAVNPHH269 pKa = 6.07LQLTLDD275 pKa = 4.35DD276 pKa = 4.3FQIPEE281 pKa = 4.49PDD283 pKa = 3.35QPLLINSFDD292 pKa = 3.89LVIHH296 pKa = 6.21GFEE299 pKa = 4.29DD300 pKa = 3.41VPYY303 pKa = 9.27ILPPEE308 pKa = 4.35PPGPQIVMVQKK319 pKa = 10.65RR320 pKa = 11.84YY321 pKa = 8.05QCPWYY326 pKa = 10.18GCRR329 pKa = 11.84TILRR333 pKa = 11.84TLVGLRR339 pKa = 11.84AHH341 pKa = 6.8IRR343 pKa = 11.84LEE345 pKa = 4.05HH346 pKa = 6.03QGRR349 pKa = 11.84PTVRR353 pKa = 11.84CPYY356 pKa = 10.33CPLKK360 pKa = 10.98YY361 pKa = 10.46GEE363 pKa = 4.95GYY365 pKa = 10.09NLHH368 pKa = 6.41RR369 pKa = 11.84HH370 pKa = 5.19VRR372 pKa = 11.84WKK374 pKa = 10.88HH375 pKa = 5.58PDD377 pKa = 3.37FLDD380 pKa = 4.28DD381 pKa = 4.56FVNFAPFF388 pKa = 3.3
MM1 pKa = 8.03GDD3 pKa = 3.49TFHH6 pKa = 7.89PDD8 pKa = 3.22NSWSLLDD15 pKa = 5.4DD16 pKa = 4.08DD17 pKa = 4.71TPAYY21 pKa = 10.43DD22 pKa = 5.15DD23 pKa = 3.99YY24 pKa = 11.88PEE26 pKa = 4.73VIPLATTEE34 pKa = 4.17YY35 pKa = 10.63VDD37 pKa = 4.1PFPAPFSTQGYY48 pKa = 8.69APRR51 pKa = 11.84EE52 pKa = 4.0IIQAVWSVSGAPSNQNIPAAAPVPNTNPDD81 pKa = 3.37PAAAPVIDD89 pKa = 4.48PNLNQTAAPTVDD101 pKa = 3.59SNLNNTEE108 pKa = 4.02APVVDD113 pKa = 4.04PGLYY117 pKa = 9.92PSVAPGTTFYY127 pKa = 10.79FDD129 pKa = 3.23QVVGVFTGSDD139 pKa = 3.98LDD141 pKa = 3.56PTMPLFLEE149 pKa = 4.88PDD151 pKa = 2.98WSFPPLIQSMPPGVSPTADD170 pKa = 3.45ATPTAAVDD178 pKa = 3.51ATMDD182 pKa = 4.0PAIDD186 pKa = 4.1PSQDD190 pKa = 3.03VAADD194 pKa = 3.67PAIDD198 pKa = 4.11PSQDD202 pKa = 3.03VAADD206 pKa = 3.64PAIDD210 pKa = 4.26PNLDD214 pKa = 3.03AAADD218 pKa = 4.0PNIDD222 pKa = 3.91PNLDD226 pKa = 3.12AAADD230 pKa = 4.08PNIHH234 pKa = 6.89SDD236 pKa = 3.99LDD238 pKa = 4.16LAALWAEE245 pKa = 4.67LDD247 pKa = 3.43QTAAPGNDD255 pKa = 3.05TGPAQTVVPAVNPHH269 pKa = 6.07LQLTLDD275 pKa = 4.35DD276 pKa = 4.3FQIPEE281 pKa = 4.49PDD283 pKa = 3.35QPLLINSFDD292 pKa = 3.89LVIHH296 pKa = 6.21GFEE299 pKa = 4.29DD300 pKa = 3.41VPYY303 pKa = 9.27ILPPEE308 pKa = 4.35PPGPQIVMVQKK319 pKa = 10.65RR320 pKa = 11.84YY321 pKa = 8.05QCPWYY326 pKa = 10.18GCRR329 pKa = 11.84TILRR333 pKa = 11.84TLVGLRR339 pKa = 11.84AHH341 pKa = 6.8IRR343 pKa = 11.84LEE345 pKa = 4.05HH346 pKa = 6.03QGRR349 pKa = 11.84PTVRR353 pKa = 11.84CPYY356 pKa = 10.33CPLKK360 pKa = 10.98YY361 pKa = 10.46GEE363 pKa = 4.95GYY365 pKa = 10.09NLHH368 pKa = 6.41RR369 pKa = 11.84HH370 pKa = 5.19VRR372 pKa = 11.84WKK374 pKa = 10.88HH375 pKa = 5.58PDD377 pKa = 3.37FLDD380 pKa = 4.28DD381 pKa = 4.56FVNFAPFF388 pKa = 3.3
Molecular weight: 41.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A319DGA6|A0A319DGA6_9EURO NIF-domain-containing protein OS=Aspergillus ellipticus CBS 707.79 OX=1448320 GN=BO71DRAFT_320935 PE=4 SV=1
MM1 pKa = 7.51SSSSQNPSPKK11 pKa = 10.32GKK13 pKa = 9.83NPKK16 pKa = 9.16NQPQTPTPTSKK27 pKa = 10.89SNPLRR32 pKa = 11.84ARR34 pKa = 11.84ARR36 pKa = 11.84ARR38 pKa = 11.84ARR40 pKa = 11.84ARR42 pKa = 11.84ARR44 pKa = 11.84VRR46 pKa = 11.84VRR48 pKa = 11.84ARR50 pKa = 11.84VRR52 pKa = 11.84VRR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84ARR59 pKa = 11.84PLRR62 pKa = 11.84NNPNNPNNPNPTPNQHH78 pKa = 6.16ATPPSPLAAFPRR90 pKa = 11.84SVTCAGAA97 pKa = 3.3
MM1 pKa = 7.51SSSSQNPSPKK11 pKa = 10.32GKK13 pKa = 9.83NPKK16 pKa = 9.16NQPQTPTPTSKK27 pKa = 10.89SNPLRR32 pKa = 11.84ARR34 pKa = 11.84ARR36 pKa = 11.84ARR38 pKa = 11.84ARR40 pKa = 11.84ARR42 pKa = 11.84ARR44 pKa = 11.84VRR46 pKa = 11.84VRR48 pKa = 11.84ARR50 pKa = 11.84VRR52 pKa = 11.84VRR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84ARR59 pKa = 11.84PLRR62 pKa = 11.84NNPNNPNNPNPTPNQHH78 pKa = 6.16ATPPSPLAAFPRR90 pKa = 11.84SVTCAGAA97 pKa = 3.3
Molecular weight: 10.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5629130 |
49 |
6272 |
437.7 |
48.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.507 ± 0.018 | 1.337 ± 0.008 |
5.549 ± 0.017 | 5.956 ± 0.021 |
3.768 ± 0.013 | 6.915 ± 0.021 |
2.49 ± 0.009 | 4.923 ± 0.016 |
4.286 ± 0.021 | 9.197 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.188 ± 0.008 | 3.493 ± 0.012 |
6.232 ± 0.026 | 3.981 ± 0.015 |
6.219 ± 0.018 | 8.327 ± 0.025 |
6.042 ± 0.016 | 6.229 ± 0.017 |
1.514 ± 0.009 | 2.847 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
