
Lucifera butyrica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Veillonellales; Veillonellaceae; Lucifera
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
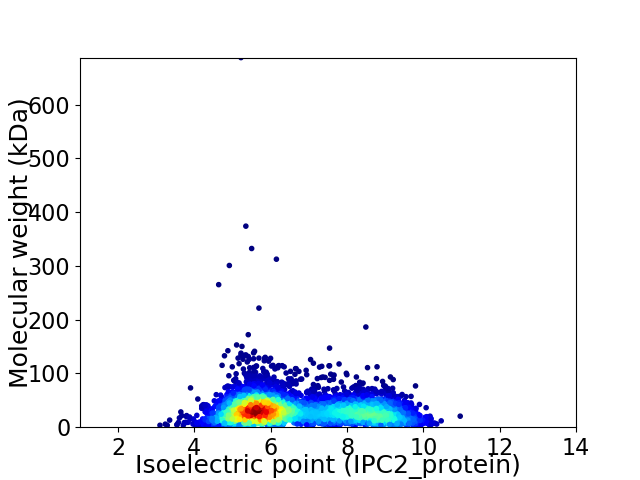
Virtual 2D-PAGE plot for 5043 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A498RH87|A0A498RH87_9FIRM AAA_31 domain-containing protein (Fragment) OS=Lucifera butyrica OX=1351585 GN=LUCI_4742 PE=4 SV=1
MM1 pKa = 7.63ASSTSSISLSTTSSSALGTTHH22 pKa = 7.87LSNLVSGLDD31 pKa = 3.39VDD33 pKa = 4.81ALVSAQILADD43 pKa = 4.52SIPLTLLQNTVAKK56 pKa = 9.18LTAQKK61 pKa = 10.77SDD63 pKa = 3.55YY64 pKa = 9.15QTLEE68 pKa = 3.79TDD70 pKa = 3.77LQTLQYY76 pKa = 11.13ALQDD80 pKa = 3.81LTYY83 pKa = 9.19STAFSTRR90 pKa = 11.84TATSSDD96 pKa = 3.58EE97 pKa = 4.3KK98 pKa = 11.27VVTASTKK105 pKa = 8.81TGASAGSYY113 pKa = 9.87SINVSQLATTSSVTGTALSSFNDD136 pKa = 3.86GTKK139 pKa = 9.54ATLTGNVSLSGYY151 pKa = 9.58NGNDD155 pKa = 3.02STALTSLGITPGTVTINGASIVITSGDD182 pKa = 3.77TINTVLNKK190 pKa = 9.94ISASSAGVTATLNSSTGDD208 pKa = 3.27VVLTQKK214 pKa = 9.32TAGSSDD220 pKa = 4.06TITLGSDD227 pKa = 3.02GGTNLFNAFGLSSGNGTLVAGQDD250 pKa = 3.28ATDD253 pKa = 3.72AQTLTQVFGSSLQAGYY269 pKa = 10.29FSINGTVFQVNPTTDD284 pKa = 3.32TLDD287 pKa = 4.2SILSEE292 pKa = 4.26INSSTTAGVQAFYY305 pKa = 11.19NSTSKK310 pKa = 9.66TVSIVSTTAGSQGSVTLGTYY330 pKa = 10.68GSGGTDD336 pKa = 2.85SSNLLDD342 pKa = 4.06LLKK345 pKa = 10.35VTTAGGNTPAVTTGTDD361 pKa = 2.85ASVTVNGVAVAANGNSITMNGNTFALQGAGTATVTVSTNYY401 pKa = 10.28DD402 pKa = 3.61AIITKK407 pKa = 8.46VQNVVTQYY415 pKa = 11.31NSVVDD420 pKa = 4.97AINAKK425 pKa = 9.43IQEE428 pKa = 4.55APSSSSDD435 pKa = 3.34SSSSASTGDD444 pKa = 3.39LHH446 pKa = 7.57LDD448 pKa = 3.46PFLEE452 pKa = 6.0GIIQDD457 pKa = 4.11LGSFSSTVLNSPNSVYY473 pKa = 9.07QTLSQVGITTGDD485 pKa = 3.2VGQSVTDD492 pKa = 3.59SEE494 pKa = 5.01LGHH497 pKa = 7.11LSLNTDD503 pKa = 3.69TLKK506 pKa = 10.04TALEE510 pKa = 4.15TNMAGVEE517 pKa = 4.27SLFGNTTVYY526 pKa = 10.69VDD528 pKa = 3.68SEE530 pKa = 4.59KK531 pKa = 11.22VGTSSSSQLTYY542 pKa = 11.28SLAHH546 pKa = 6.64GDD548 pKa = 3.15ITGTPTVMVGSKK560 pKa = 10.1QYY562 pKa = 9.92TVVTAAQLSTDD573 pKa = 3.67DD574 pKa = 4.1TNAKK578 pKa = 10.15NSSTTIYY585 pKa = 9.64EE586 pKa = 4.29CSVDD590 pKa = 3.34ATTGKK595 pKa = 8.34ITFATGAGTFPNGSQITVSYY615 pKa = 10.71NYY617 pKa = 10.17TNTTGGSGTGIFSQMQAFINTYY639 pKa = 5.84TTVGGQFDD647 pKa = 4.12TMIGSNGSLTDD658 pKa = 3.88TISYY662 pKa = 11.19DD663 pKa = 3.41NDD665 pKa = 3.45RR666 pKa = 11.84VSEE669 pKa = 4.08MQDD672 pKa = 3.13RR673 pKa = 11.84INNEE677 pKa = 3.5QSSLYY682 pKa = 9.91TYY684 pKa = 9.71YY685 pKa = 11.59NNMLSQLQTLSSEE698 pKa = 4.08NTMVSALLSGLSSSSSSSSKK718 pKa = 10.91GG719 pKa = 3.27
MM1 pKa = 7.63ASSTSSISLSTTSSSALGTTHH22 pKa = 7.87LSNLVSGLDD31 pKa = 3.39VDD33 pKa = 4.81ALVSAQILADD43 pKa = 4.52SIPLTLLQNTVAKK56 pKa = 9.18LTAQKK61 pKa = 10.77SDD63 pKa = 3.55YY64 pKa = 9.15QTLEE68 pKa = 3.79TDD70 pKa = 3.77LQTLQYY76 pKa = 11.13ALQDD80 pKa = 3.81LTYY83 pKa = 9.19STAFSTRR90 pKa = 11.84TATSSDD96 pKa = 3.58EE97 pKa = 4.3KK98 pKa = 11.27VVTASTKK105 pKa = 8.81TGASAGSYY113 pKa = 9.87SINVSQLATTSSVTGTALSSFNDD136 pKa = 3.86GTKK139 pKa = 9.54ATLTGNVSLSGYY151 pKa = 9.58NGNDD155 pKa = 3.02STALTSLGITPGTVTINGASIVITSGDD182 pKa = 3.77TINTVLNKK190 pKa = 9.94ISASSAGVTATLNSSTGDD208 pKa = 3.27VVLTQKK214 pKa = 9.32TAGSSDD220 pKa = 4.06TITLGSDD227 pKa = 3.02GGTNLFNAFGLSSGNGTLVAGQDD250 pKa = 3.28ATDD253 pKa = 3.72AQTLTQVFGSSLQAGYY269 pKa = 10.29FSINGTVFQVNPTTDD284 pKa = 3.32TLDD287 pKa = 4.2SILSEE292 pKa = 4.26INSSTTAGVQAFYY305 pKa = 11.19NSTSKK310 pKa = 9.66TVSIVSTTAGSQGSVTLGTYY330 pKa = 10.68GSGGTDD336 pKa = 2.85SSNLLDD342 pKa = 4.06LLKK345 pKa = 10.35VTTAGGNTPAVTTGTDD361 pKa = 2.85ASVTVNGVAVAANGNSITMNGNTFALQGAGTATVTVSTNYY401 pKa = 10.28DD402 pKa = 3.61AIITKK407 pKa = 8.46VQNVVTQYY415 pKa = 11.31NSVVDD420 pKa = 4.97AINAKK425 pKa = 9.43IQEE428 pKa = 4.55APSSSSDD435 pKa = 3.34SSSSASTGDD444 pKa = 3.39LHH446 pKa = 7.57LDD448 pKa = 3.46PFLEE452 pKa = 6.0GIIQDD457 pKa = 4.11LGSFSSTVLNSPNSVYY473 pKa = 9.07QTLSQVGITTGDD485 pKa = 3.2VGQSVTDD492 pKa = 3.59SEE494 pKa = 5.01LGHH497 pKa = 7.11LSLNTDD503 pKa = 3.69TLKK506 pKa = 10.04TALEE510 pKa = 4.15TNMAGVEE517 pKa = 4.27SLFGNTTVYY526 pKa = 10.69VDD528 pKa = 3.68SEE530 pKa = 4.59KK531 pKa = 11.22VGTSSSSQLTYY542 pKa = 11.28SLAHH546 pKa = 6.64GDD548 pKa = 3.15ITGTPTVMVGSKK560 pKa = 10.1QYY562 pKa = 9.92TVVTAAQLSTDD573 pKa = 3.67DD574 pKa = 4.1TNAKK578 pKa = 10.15NSSTTIYY585 pKa = 9.64EE586 pKa = 4.29CSVDD590 pKa = 3.34ATTGKK595 pKa = 8.34ITFATGAGTFPNGSQITVSYY615 pKa = 10.71NYY617 pKa = 10.17TNTTGGSGTGIFSQMQAFINTYY639 pKa = 5.84TTVGGQFDD647 pKa = 4.12TMIGSNGSLTDD658 pKa = 3.88TISYY662 pKa = 11.19DD663 pKa = 3.41NDD665 pKa = 3.45RR666 pKa = 11.84VSEE669 pKa = 4.08MQDD672 pKa = 3.13RR673 pKa = 11.84INNEE677 pKa = 3.5QSSLYY682 pKa = 9.91TYY684 pKa = 9.71YY685 pKa = 11.59NNMLSQLQTLSSEE698 pKa = 4.08NTMVSALLSGLSSSSSSSSKK718 pKa = 10.91GG719 pKa = 3.27
Molecular weight: 73.09 kDa
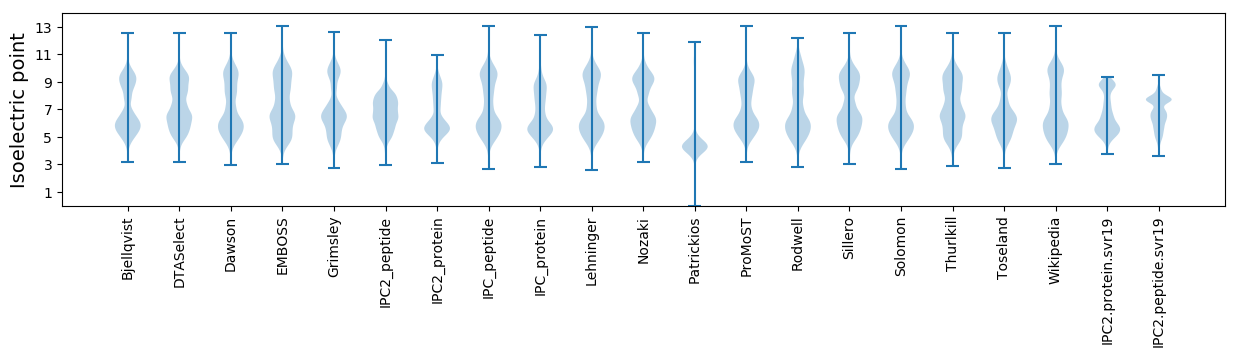
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A498R6D0|A0A498R6D0_9FIRM HD-GYP domain-containing protein OS=Lucifera butyrica OX=1351585 GN=LUCI_1975 PE=4 SV=1
MM1 pKa = 7.58TIDD4 pKa = 3.21KK5 pKa = 10.6NSRR8 pKa = 11.84RR9 pKa = 11.84NSRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84LATRR18 pKa = 11.84TAIHH22 pKa = 6.31FVLPFAIQTAIHH34 pKa = 6.34FVLPLARR41 pKa = 11.84LPALPLLRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84LAILTAGHH59 pKa = 6.35GRR61 pKa = 11.84FFATPAAASHH71 pKa = 5.77GRR73 pKa = 11.84FLAFLTAQIAARR85 pKa = 11.84APGTKK90 pKa = 9.45TNFVRR95 pKa = 11.84KK96 pKa = 9.46YY97 pKa = 9.77RR98 pKa = 11.84NQSNRR103 pKa = 11.84KK104 pKa = 6.13TADD107 pKa = 3.01AVFLLDD113 pKa = 4.6SYY115 pKa = 11.41SGHH118 pKa = 6.26NPGAASSLQRR128 pKa = 11.84FLRR131 pKa = 11.84LSQRR135 pKa = 11.84GNQRR139 pKa = 11.84LPSRR143 pKa = 11.84FGRR146 pKa = 11.84HH147 pKa = 4.15KK148 pKa = 9.75LHH150 pKa = 6.99RR151 pKa = 11.84RR152 pKa = 11.84FHH154 pKa = 6.03LRR156 pKa = 11.84QHH158 pKa = 6.37AAGSKK163 pKa = 10.16LPLRR167 pKa = 11.84NIFFRR172 pKa = 11.84FFQTHH177 pKa = 5.29LRR179 pKa = 3.87
MM1 pKa = 7.58TIDD4 pKa = 3.21KK5 pKa = 10.6NSRR8 pKa = 11.84RR9 pKa = 11.84NSRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84LATRR18 pKa = 11.84TAIHH22 pKa = 6.31FVLPFAIQTAIHH34 pKa = 6.34FVLPLARR41 pKa = 11.84LPALPLLRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84LAILTAGHH59 pKa = 6.35GRR61 pKa = 11.84FFATPAAASHH71 pKa = 5.77GRR73 pKa = 11.84FLAFLTAQIAARR85 pKa = 11.84APGTKK90 pKa = 9.45TNFVRR95 pKa = 11.84KK96 pKa = 9.46YY97 pKa = 9.77RR98 pKa = 11.84NQSNRR103 pKa = 11.84KK104 pKa = 6.13TADD107 pKa = 3.01AVFLLDD113 pKa = 4.6SYY115 pKa = 11.41SGHH118 pKa = 6.26NPGAASSLQRR128 pKa = 11.84FLRR131 pKa = 11.84LSQRR135 pKa = 11.84GNQRR139 pKa = 11.84LPSRR143 pKa = 11.84FGRR146 pKa = 11.84HH147 pKa = 4.15KK148 pKa = 9.75LHH150 pKa = 6.99RR151 pKa = 11.84RR152 pKa = 11.84FHH154 pKa = 6.03LRR156 pKa = 11.84QHH158 pKa = 6.37AAGSKK163 pKa = 10.16LPLRR167 pKa = 11.84NIFFRR172 pKa = 11.84FFQTHH177 pKa = 5.29LRR179 pKa = 3.87
Molecular weight: 20.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1511077 |
23 |
6215 |
299.6 |
33.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.146 ± 0.046 | 1.215 ± 0.016 |
4.789 ± 0.026 | 6.168 ± 0.036 |
4.037 ± 0.028 | 7.654 ± 0.04 |
1.881 ± 0.016 | 7.217 ± 0.033 |
5.643 ± 0.034 | 9.99 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.738 ± 0.016 | 4.06 ± 0.026 |
4.066 ± 0.023 | 3.849 ± 0.023 |
4.965 ± 0.032 | 5.391 ± 0.025 |
5.377 ± 0.027 | 7.397 ± 0.033 |
1.05 ± 0.014 | 3.367 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
