
Hubei sobemo-like virus 30
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
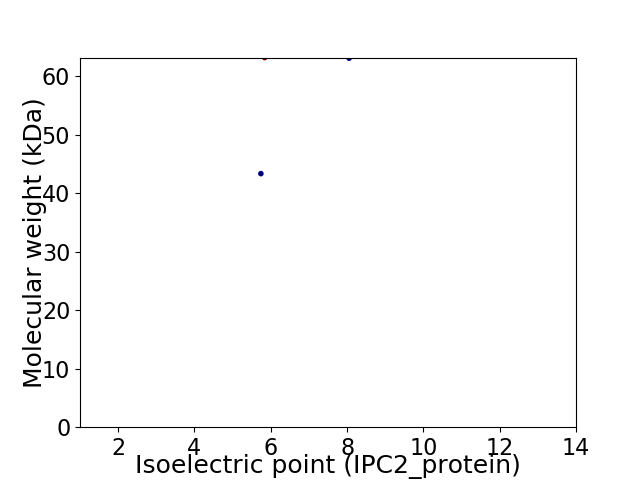
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEK2|A0A1L3KEK2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 30 OX=1923217 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.25NLLSAFEE9 pKa = 4.16SVKK12 pKa = 10.1WSIRR16 pKa = 11.84KK17 pKa = 9.44DD18 pKa = 3.58FLSFDD23 pKa = 3.63HH24 pKa = 6.96FSDD27 pKa = 3.7VVRR30 pKa = 11.84NKK32 pKa = 10.12IEE34 pKa = 4.03WTSSPGYY41 pKa = 9.6PYY43 pKa = 11.29LLRR46 pKa = 11.84ATSNGIYY53 pKa = 10.37FNVKK57 pKa = 10.29NGIPDD62 pKa = 3.49EE63 pKa = 4.17NRR65 pKa = 11.84LLEE68 pKa = 4.29VYY70 pKa = 9.82QVVMSQINDD79 pKa = 3.42RR80 pKa = 11.84SSDD83 pKa = 4.0PIRR86 pKa = 11.84LFIKK90 pKa = 10.24QEE92 pKa = 3.93PLKK95 pKa = 10.82HH96 pKa = 6.03EE97 pKa = 4.45KK98 pKa = 10.31LDD100 pKa = 3.45SGRR103 pKa = 11.84YY104 pKa = 8.73RR105 pKa = 11.84LISSVSIIDD114 pKa = 3.68QIIDD118 pKa = 3.27HH119 pKa = 6.32MLFDD123 pKa = 5.63DD124 pKa = 4.65MNDD127 pKa = 3.73MITDD131 pKa = 3.2DD132 pKa = 3.78WMFVPPKK139 pKa = 9.8IGWSAFTGGWKK150 pKa = 10.44FMPKK154 pKa = 10.32LSEE157 pKa = 4.17QPWALDD163 pKa = 3.22KK164 pKa = 11.54SSWDD168 pKa = 3.06WTVNLWLFDD177 pKa = 3.27IVLDD181 pKa = 5.22MRR183 pKa = 11.84IQLCKK188 pKa = 9.96NLSRR192 pKa = 11.84EE193 pKa = 4.21WVDD196 pKa = 3.36LALWRR201 pKa = 11.84YY202 pKa = 7.49SQLYY206 pKa = 9.27IEE208 pKa = 5.55PIFVTTGGLLLKK220 pKa = 10.49QITPGVMKK228 pKa = 10.11SGCVNTIVDD237 pKa = 3.84NSIMQVILHH246 pKa = 6.11LRR248 pKa = 11.84VCFEE252 pKa = 3.83EE253 pKa = 5.66GIRR256 pKa = 11.84PDD258 pKa = 3.14WLFSLGDD265 pKa = 3.49DD266 pKa = 3.9TLQKK270 pKa = 10.68KK271 pKa = 8.5PAEE274 pKa = 4.03PEE276 pKa = 3.72RR277 pKa = 11.84YY278 pKa = 8.76LRR280 pKa = 11.84AMSKK284 pKa = 10.19YY285 pKa = 10.26AIVKK289 pKa = 8.59QAHH292 pKa = 6.82DD293 pKa = 3.59VTEE296 pKa = 4.47FAGHH300 pKa = 6.63RR301 pKa = 11.84FDD303 pKa = 5.59GIHH306 pKa = 5.82VEE308 pKa = 3.64PVYY311 pKa = 10.36RR312 pKa = 11.84SKK314 pKa = 10.96HH315 pKa = 5.35SYY317 pKa = 10.05ILLHH321 pKa = 6.3MDD323 pKa = 4.16PANLEE328 pKa = 3.99AMANSYY334 pKa = 10.69CLLYY338 pKa = 10.36HH339 pKa = 6.49RR340 pKa = 11.84SVYY343 pKa = 10.47RR344 pKa = 11.84DD345 pKa = 3.31VIRR348 pKa = 11.84DD349 pKa = 3.71LFAQMGAKK357 pKa = 9.89LPNLRR362 pKa = 11.84EE363 pKa = 3.81LDD365 pKa = 4.36LIFDD369 pKa = 4.42GMM371 pKa = 4.03
MM1 pKa = 7.59KK2 pKa = 10.25NLLSAFEE9 pKa = 4.16SVKK12 pKa = 10.1WSIRR16 pKa = 11.84KK17 pKa = 9.44DD18 pKa = 3.58FLSFDD23 pKa = 3.63HH24 pKa = 6.96FSDD27 pKa = 3.7VVRR30 pKa = 11.84NKK32 pKa = 10.12IEE34 pKa = 4.03WTSSPGYY41 pKa = 9.6PYY43 pKa = 11.29LLRR46 pKa = 11.84ATSNGIYY53 pKa = 10.37FNVKK57 pKa = 10.29NGIPDD62 pKa = 3.49EE63 pKa = 4.17NRR65 pKa = 11.84LLEE68 pKa = 4.29VYY70 pKa = 9.82QVVMSQINDD79 pKa = 3.42RR80 pKa = 11.84SSDD83 pKa = 4.0PIRR86 pKa = 11.84LFIKK90 pKa = 10.24QEE92 pKa = 3.93PLKK95 pKa = 10.82HH96 pKa = 6.03EE97 pKa = 4.45KK98 pKa = 10.31LDD100 pKa = 3.45SGRR103 pKa = 11.84YY104 pKa = 8.73RR105 pKa = 11.84LISSVSIIDD114 pKa = 3.68QIIDD118 pKa = 3.27HH119 pKa = 6.32MLFDD123 pKa = 5.63DD124 pKa = 4.65MNDD127 pKa = 3.73MITDD131 pKa = 3.2DD132 pKa = 3.78WMFVPPKK139 pKa = 9.8IGWSAFTGGWKK150 pKa = 10.44FMPKK154 pKa = 10.32LSEE157 pKa = 4.17QPWALDD163 pKa = 3.22KK164 pKa = 11.54SSWDD168 pKa = 3.06WTVNLWLFDD177 pKa = 3.27IVLDD181 pKa = 5.22MRR183 pKa = 11.84IQLCKK188 pKa = 9.96NLSRR192 pKa = 11.84EE193 pKa = 4.21WVDD196 pKa = 3.36LALWRR201 pKa = 11.84YY202 pKa = 7.49SQLYY206 pKa = 9.27IEE208 pKa = 5.55PIFVTTGGLLLKK220 pKa = 10.49QITPGVMKK228 pKa = 10.11SGCVNTIVDD237 pKa = 3.84NSIMQVILHH246 pKa = 6.11LRR248 pKa = 11.84VCFEE252 pKa = 3.83EE253 pKa = 5.66GIRR256 pKa = 11.84PDD258 pKa = 3.14WLFSLGDD265 pKa = 3.49DD266 pKa = 3.9TLQKK270 pKa = 10.68KK271 pKa = 8.5PAEE274 pKa = 4.03PEE276 pKa = 3.72RR277 pKa = 11.84YY278 pKa = 8.76LRR280 pKa = 11.84AMSKK284 pKa = 10.19YY285 pKa = 10.26AIVKK289 pKa = 8.59QAHH292 pKa = 6.82DD293 pKa = 3.59VTEE296 pKa = 4.47FAGHH300 pKa = 6.63RR301 pKa = 11.84FDD303 pKa = 5.59GIHH306 pKa = 5.82VEE308 pKa = 3.64PVYY311 pKa = 10.36RR312 pKa = 11.84SKK314 pKa = 10.96HH315 pKa = 5.35SYY317 pKa = 10.05ILLHH321 pKa = 6.3MDD323 pKa = 4.16PANLEE328 pKa = 3.99AMANSYY334 pKa = 10.69CLLYY338 pKa = 10.36HH339 pKa = 6.49RR340 pKa = 11.84SVYY343 pKa = 10.47RR344 pKa = 11.84DD345 pKa = 3.31VIRR348 pKa = 11.84DD349 pKa = 3.71LFAQMGAKK357 pKa = 9.89LPNLRR362 pKa = 11.84EE363 pKa = 3.81LDD365 pKa = 4.36LIFDD369 pKa = 4.42GMM371 pKa = 4.03
Molecular weight: 43.32 kDa
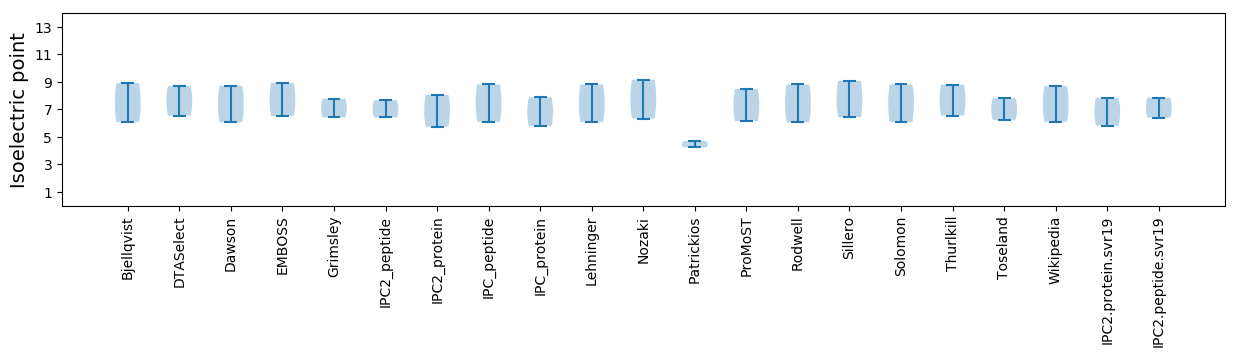
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEK2|A0A1L3KEK2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 30 OX=1923217 PE=4 SV=1
MM1 pKa = 7.43TFLLVSFLVRR11 pKa = 11.84WAKK14 pKa = 10.4HH15 pKa = 3.99INARR19 pKa = 11.84VCALVSAGSEE29 pKa = 3.97GLYY32 pKa = 10.64SVLDD36 pKa = 4.77EE37 pKa = 4.87ISPSPTPEE45 pKa = 3.09IRR47 pKa = 11.84KK48 pKa = 8.47QKK50 pKa = 9.48KK51 pKa = 9.52GKK53 pKa = 9.55VLTVIDD59 pKa = 5.18DD60 pKa = 3.48ILRR63 pKa = 11.84NIDD66 pKa = 3.12FRR68 pKa = 11.84EE69 pKa = 3.88YY70 pKa = 10.54MDD72 pKa = 5.18FDD74 pKa = 4.42WIDD77 pKa = 3.59ALVIAACSVLTLWASYY93 pKa = 9.73MIFRR97 pKa = 11.84VMGRR101 pKa = 11.84NGRR104 pKa = 11.84RR105 pKa = 11.84IVQRR109 pKa = 11.84LRR111 pKa = 11.84GIQNEE116 pKa = 3.81AMIDD120 pKa = 3.63GSAFHH125 pKa = 6.99AGEE128 pKa = 3.93IPKK131 pKa = 9.66FQIGIARR138 pKa = 11.84SKK140 pKa = 10.91LFVDD144 pKa = 3.08VHH146 pKa = 7.42VGFGIRR152 pKa = 11.84SKK154 pKa = 11.34NFLITPRR161 pKa = 11.84HH162 pKa = 4.83VLEE165 pKa = 4.66GVDD168 pKa = 3.81LKK170 pKa = 11.17EE171 pKa = 4.05VLLVGPTGKK180 pKa = 10.6VIVSLCPQPSRR191 pKa = 11.84LVSDD195 pKa = 3.57LTYY198 pKa = 10.39TYY200 pKa = 10.1IEE202 pKa = 4.1SAIWTKK208 pKa = 10.95LGARR212 pKa = 11.84SANLNNTPITTMVEE226 pKa = 4.22CCGKK230 pKa = 10.25DD231 pKa = 3.17GKK233 pKa = 11.49SNGRR237 pKa = 11.84IRR239 pKa = 11.84KK240 pKa = 7.22TNMFGLISYY249 pKa = 7.62TGSTLPGMSGAAYY262 pKa = 10.16VFNNQVQGIHH272 pKa = 6.42KK273 pKa = 9.88GAAGRR278 pKa = 11.84YY279 pKa = 7.84NLGVSSAVLAAEE291 pKa = 4.6LSKK294 pKa = 11.01LCPPTADD301 pKa = 3.29EE302 pKa = 4.84SLPAKK307 pKa = 10.33DD308 pKa = 3.79AEE310 pKa = 4.6DD311 pKa = 3.63YY312 pKa = 11.42VEE314 pKa = 5.78QFQQQNTWTTQQLEE328 pKa = 4.35KK329 pKa = 10.47KK330 pKa = 8.27IQEE333 pKa = 3.85MDD335 pKa = 3.68KK336 pKa = 8.9YY337 pKa = 8.97TGWSTQEE344 pKa = 3.85TMDD347 pKa = 5.09FNQTLNWDD355 pKa = 3.7EE356 pKa = 4.69SSPSTSDD363 pKa = 3.3STSGAATKK371 pKa = 10.45SNPMFVNYY379 pKa = 9.17VAQGILSDD387 pKa = 4.38CKK389 pKa = 10.81DD390 pKa = 3.16IPTVEE395 pKa = 4.16FTSRR399 pKa = 11.84LEE401 pKa = 4.34SLWSQYY407 pKa = 10.55PVILNRR413 pKa = 11.84LCAVEE418 pKa = 5.06KK419 pKa = 10.36EE420 pKa = 4.25LEE422 pKa = 4.44AIKK425 pKa = 10.62APKK428 pKa = 9.33PVANCEE434 pKa = 4.35SQTEE438 pKa = 4.28PEE440 pKa = 4.47VVPEE444 pKa = 3.87ASKK447 pKa = 11.23VVTVVDD453 pKa = 3.48KK454 pKa = 11.17TPFCFLCSKK463 pKa = 10.68HH464 pKa = 5.95FATEE468 pKa = 3.11NGYY471 pKa = 7.57QHH473 pKa = 7.18HH474 pKa = 6.99ISNSKK479 pKa = 8.92KK480 pKa = 8.29HH481 pKa = 5.22TVAEE485 pKa = 4.26SAIPGDD491 pKa = 3.74TGDD494 pKa = 3.28KK495 pKa = 10.43GKK497 pKa = 10.51VVRR500 pKa = 11.84TAGPAPFLGPATTSQKK516 pKa = 10.31RR517 pKa = 11.84KK518 pKa = 8.04KK519 pKa = 8.31TSSSKK524 pKa = 10.41SSGRR528 pKa = 11.84SARR531 pKa = 11.84RR532 pKa = 11.84NQSPPNQGHH541 pKa = 6.75PSEE544 pKa = 4.71MMSSLKK550 pKa = 10.78NIEE553 pKa = 4.54LVLKK557 pKa = 10.88NLVQVMDD564 pKa = 4.08GQKK567 pKa = 10.19SGKK570 pKa = 8.58EE571 pKa = 3.9RR572 pKa = 11.84NN573 pKa = 3.51
MM1 pKa = 7.43TFLLVSFLVRR11 pKa = 11.84WAKK14 pKa = 10.4HH15 pKa = 3.99INARR19 pKa = 11.84VCALVSAGSEE29 pKa = 3.97GLYY32 pKa = 10.64SVLDD36 pKa = 4.77EE37 pKa = 4.87ISPSPTPEE45 pKa = 3.09IRR47 pKa = 11.84KK48 pKa = 8.47QKK50 pKa = 9.48KK51 pKa = 9.52GKK53 pKa = 9.55VLTVIDD59 pKa = 5.18DD60 pKa = 3.48ILRR63 pKa = 11.84NIDD66 pKa = 3.12FRR68 pKa = 11.84EE69 pKa = 3.88YY70 pKa = 10.54MDD72 pKa = 5.18FDD74 pKa = 4.42WIDD77 pKa = 3.59ALVIAACSVLTLWASYY93 pKa = 9.73MIFRR97 pKa = 11.84VMGRR101 pKa = 11.84NGRR104 pKa = 11.84RR105 pKa = 11.84IVQRR109 pKa = 11.84LRR111 pKa = 11.84GIQNEE116 pKa = 3.81AMIDD120 pKa = 3.63GSAFHH125 pKa = 6.99AGEE128 pKa = 3.93IPKK131 pKa = 9.66FQIGIARR138 pKa = 11.84SKK140 pKa = 10.91LFVDD144 pKa = 3.08VHH146 pKa = 7.42VGFGIRR152 pKa = 11.84SKK154 pKa = 11.34NFLITPRR161 pKa = 11.84HH162 pKa = 4.83VLEE165 pKa = 4.66GVDD168 pKa = 3.81LKK170 pKa = 11.17EE171 pKa = 4.05VLLVGPTGKK180 pKa = 10.6VIVSLCPQPSRR191 pKa = 11.84LVSDD195 pKa = 3.57LTYY198 pKa = 10.39TYY200 pKa = 10.1IEE202 pKa = 4.1SAIWTKK208 pKa = 10.95LGARR212 pKa = 11.84SANLNNTPITTMVEE226 pKa = 4.22CCGKK230 pKa = 10.25DD231 pKa = 3.17GKK233 pKa = 11.49SNGRR237 pKa = 11.84IRR239 pKa = 11.84KK240 pKa = 7.22TNMFGLISYY249 pKa = 7.62TGSTLPGMSGAAYY262 pKa = 10.16VFNNQVQGIHH272 pKa = 6.42KK273 pKa = 9.88GAAGRR278 pKa = 11.84YY279 pKa = 7.84NLGVSSAVLAAEE291 pKa = 4.6LSKK294 pKa = 11.01LCPPTADD301 pKa = 3.29EE302 pKa = 4.84SLPAKK307 pKa = 10.33DD308 pKa = 3.79AEE310 pKa = 4.6DD311 pKa = 3.63YY312 pKa = 11.42VEE314 pKa = 5.78QFQQQNTWTTQQLEE328 pKa = 4.35KK329 pKa = 10.47KK330 pKa = 8.27IQEE333 pKa = 3.85MDD335 pKa = 3.68KK336 pKa = 8.9YY337 pKa = 8.97TGWSTQEE344 pKa = 3.85TMDD347 pKa = 5.09FNQTLNWDD355 pKa = 3.7EE356 pKa = 4.69SSPSTSDD363 pKa = 3.3STSGAATKK371 pKa = 10.45SNPMFVNYY379 pKa = 9.17VAQGILSDD387 pKa = 4.38CKK389 pKa = 10.81DD390 pKa = 3.16IPTVEE395 pKa = 4.16FTSRR399 pKa = 11.84LEE401 pKa = 4.34SLWSQYY407 pKa = 10.55PVILNRR413 pKa = 11.84LCAVEE418 pKa = 5.06KK419 pKa = 10.36EE420 pKa = 4.25LEE422 pKa = 4.44AIKK425 pKa = 10.62APKK428 pKa = 9.33PVANCEE434 pKa = 4.35SQTEE438 pKa = 4.28PEE440 pKa = 4.47VVPEE444 pKa = 3.87ASKK447 pKa = 11.23VVTVVDD453 pKa = 3.48KK454 pKa = 11.17TPFCFLCSKK463 pKa = 10.68HH464 pKa = 5.95FATEE468 pKa = 3.11NGYY471 pKa = 7.57QHH473 pKa = 7.18HH474 pKa = 6.99ISNSKK479 pKa = 8.92KK480 pKa = 8.29HH481 pKa = 5.22TVAEE485 pKa = 4.26SAIPGDD491 pKa = 3.74TGDD494 pKa = 3.28KK495 pKa = 10.43GKK497 pKa = 10.51VVRR500 pKa = 11.84TAGPAPFLGPATTSQKK516 pKa = 10.31RR517 pKa = 11.84KK518 pKa = 8.04KK519 pKa = 8.31TSSSKK524 pKa = 10.41SSGRR528 pKa = 11.84SARR531 pKa = 11.84RR532 pKa = 11.84NQSPPNQGHH541 pKa = 6.75PSEE544 pKa = 4.71MMSSLKK550 pKa = 10.78NIEE553 pKa = 4.54LVLKK557 pKa = 10.88NLVQVMDD564 pKa = 4.08GQKK567 pKa = 10.19SGKK570 pKa = 8.58EE571 pKa = 3.9RR572 pKa = 11.84NN573 pKa = 3.51
Molecular weight: 63.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
944 |
371 |
573 |
472.0 |
53.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.826 ± 1.082 | 1.589 ± 0.31 |
5.826 ± 1.371 | 5.191 ± 0.369 |
4.025 ± 0.501 | 5.932 ± 0.819 |
2.119 ± 0.35 | 6.356 ± 0.723 |
6.462 ± 0.486 | 9.004 ± 1.242 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.072 ± 0.589 | 4.449 ± 0.246 |
4.873 ± 0.176 | 3.919 ± 0.416 |
5.085 ± 0.349 | 8.792 ± 0.592 |
5.191 ± 1.35 | 7.309 ± 0.51 |
2.119 ± 0.677 | 2.86 ± 0.554 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
