
Hubei tombus-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.58
Get precalculated fractions of proteins
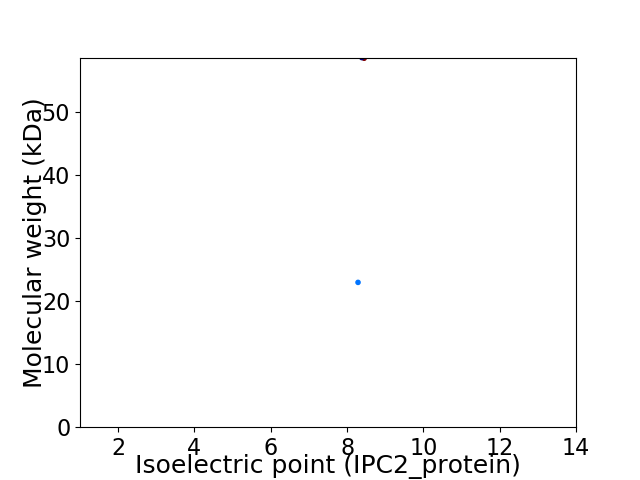
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KGL0|A0A1L3KGL0_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 5 OX=1923292 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.26SVAPDD7 pKa = 3.46HH8 pKa = 7.56PDD10 pKa = 3.0LQVRR14 pKa = 11.84KK15 pKa = 9.32RR16 pKa = 11.84WGDD19 pKa = 3.28TKK21 pKa = 10.64VRR23 pKa = 11.84KK24 pKa = 6.48TARR27 pKa = 11.84VSGISPRR34 pKa = 11.84MEE36 pKa = 3.5LKK38 pKa = 10.78SFNNDD43 pKa = 1.84IDD45 pKa = 3.97TLEE48 pKa = 4.21RR49 pKa = 11.84AVKK52 pKa = 10.09EE53 pKa = 3.55RR54 pKa = 11.84VFFVKK59 pKa = 10.48NDD61 pKa = 3.27LGEE64 pKa = 4.22FVEE67 pKa = 4.87PPKK70 pKa = 10.08PAPHH74 pKa = 6.96HH75 pKa = 5.69FRR77 pKa = 11.84KK78 pKa = 10.43AMLATEE84 pKa = 4.11KK85 pKa = 10.92LLVRR89 pKa = 11.84RR90 pKa = 11.84LPKK93 pKa = 9.48TAPLSRR99 pKa = 11.84DD100 pKa = 3.05AFVEE104 pKa = 4.07TFRR107 pKa = 11.84GRR109 pKa = 11.84KK110 pKa = 8.6RR111 pKa = 11.84EE112 pKa = 3.9IYY114 pKa = 10.09AKK116 pKa = 10.2AVEE119 pKa = 4.45SLSSQSVTQRR129 pKa = 11.84DD130 pKa = 3.11AGVQVFVKK138 pKa = 10.42FEE140 pKa = 3.92KK141 pKa = 10.02TDD143 pKa = 3.46YY144 pKa = 9.79TRR146 pKa = 11.84KK147 pKa = 9.53KK148 pKa = 10.49DD149 pKa = 3.43PVPRR153 pKa = 11.84VISPRR158 pKa = 11.84NPRR161 pKa = 11.84YY162 pKa = 8.55NVEE165 pKa = 3.58VGRR168 pKa = 11.84YY169 pKa = 8.5LRR171 pKa = 11.84TIEE174 pKa = 4.26EE175 pKa = 4.41PLFHH179 pKa = 7.06SLSQLFEE186 pKa = 4.11GKK188 pKa = 8.43RR189 pKa = 11.84TVFKK193 pKa = 10.91GMNAADD199 pKa = 4.42SGRR202 pKa = 11.84AMYY205 pKa = 10.08DD206 pKa = 2.66LWSSFRR212 pKa = 11.84KK213 pKa = 9.13PVAVGLDD220 pKa = 3.18ASRR223 pKa = 11.84FDD225 pKa = 3.56QHH227 pKa = 6.64VSKK230 pKa = 10.97CALQWEE236 pKa = 4.6HH237 pKa = 7.14AIYY240 pKa = 8.55PQCFTSSGDD249 pKa = 3.33RR250 pKa = 11.84AEE252 pKa = 4.61LRR254 pKa = 11.84RR255 pKa = 11.84LLKK258 pKa = 9.72WQVHH262 pKa = 5.09NKK264 pKa = 9.46CVGYY268 pKa = 10.33CADD271 pKa = 3.94GRR273 pKa = 11.84IKK275 pKa = 9.36YY276 pKa = 7.66TKK278 pKa = 9.85EE279 pKa = 3.56GTRR282 pKa = 11.84MSGDD286 pKa = 3.41MNTSLGNCVLMCSMIKK302 pKa = 10.05QYY304 pKa = 11.37SLDD307 pKa = 3.57RR308 pKa = 11.84GVRR311 pKa = 11.84TLLANNGDD319 pKa = 3.82DD320 pKa = 3.43CVVFMEE326 pKa = 4.63ADD328 pKa = 3.23EE329 pKa = 4.4LARR332 pKa = 11.84FSSGLDD338 pKa = 2.85EE339 pKa = 4.68WFRR342 pKa = 11.84AMGFNMVVEE351 pKa = 4.57PPCYY355 pKa = 9.21QFEE358 pKa = 4.42EE359 pKa = 4.53IEE361 pKa = 4.43FCQTHH366 pKa = 7.26PIYY369 pKa = 10.74VGPKK373 pKa = 8.16HH374 pKa = 6.94SDD376 pKa = 3.25YY377 pKa = 11.69LMVRR381 pKa = 11.84HH382 pKa = 6.27PKK384 pKa = 8.62WALAKK389 pKa = 10.66DD390 pKa = 3.77SMCIHH395 pKa = 7.12GFPTSKK401 pKa = 9.78IYY403 pKa = 10.36KK404 pKa = 8.74AWLDD408 pKa = 3.6AVGTGGLAMTGGVPIFQDD426 pKa = 5.37FYY428 pKa = 11.37STYY431 pKa = 9.9CKK433 pKa = 10.56YY434 pKa = 10.33GVRR437 pKa = 11.84GKK439 pKa = 7.3THH441 pKa = 6.07FHH443 pKa = 5.02EE444 pKa = 4.59QSWGVRR450 pKa = 11.84SLQQGMVRR458 pKa = 11.84KK459 pKa = 9.41YY460 pKa = 10.73GAVLPEE466 pKa = 4.21TRR468 pKa = 11.84ASFYY472 pKa = 10.04WAFGVLPDD480 pKa = 3.91EE481 pKa = 4.27QLAIEE486 pKa = 4.25DD487 pKa = 4.68FYY489 pKa = 11.64RR490 pKa = 11.84GVKK493 pKa = 9.97LDD495 pKa = 3.7EE496 pKa = 4.14AWRR499 pKa = 11.84DD500 pKa = 3.51EE501 pKa = 5.94LEE503 pKa = 4.25FQPLLPLL510 pKa = 4.0
MM1 pKa = 7.44KK2 pKa = 10.26SVAPDD7 pKa = 3.46HH8 pKa = 7.56PDD10 pKa = 3.0LQVRR14 pKa = 11.84KK15 pKa = 9.32RR16 pKa = 11.84WGDD19 pKa = 3.28TKK21 pKa = 10.64VRR23 pKa = 11.84KK24 pKa = 6.48TARR27 pKa = 11.84VSGISPRR34 pKa = 11.84MEE36 pKa = 3.5LKK38 pKa = 10.78SFNNDD43 pKa = 1.84IDD45 pKa = 3.97TLEE48 pKa = 4.21RR49 pKa = 11.84AVKK52 pKa = 10.09EE53 pKa = 3.55RR54 pKa = 11.84VFFVKK59 pKa = 10.48NDD61 pKa = 3.27LGEE64 pKa = 4.22FVEE67 pKa = 4.87PPKK70 pKa = 10.08PAPHH74 pKa = 6.96HH75 pKa = 5.69FRR77 pKa = 11.84KK78 pKa = 10.43AMLATEE84 pKa = 4.11KK85 pKa = 10.92LLVRR89 pKa = 11.84RR90 pKa = 11.84LPKK93 pKa = 9.48TAPLSRR99 pKa = 11.84DD100 pKa = 3.05AFVEE104 pKa = 4.07TFRR107 pKa = 11.84GRR109 pKa = 11.84KK110 pKa = 8.6RR111 pKa = 11.84EE112 pKa = 3.9IYY114 pKa = 10.09AKK116 pKa = 10.2AVEE119 pKa = 4.45SLSSQSVTQRR129 pKa = 11.84DD130 pKa = 3.11AGVQVFVKK138 pKa = 10.42FEE140 pKa = 3.92KK141 pKa = 10.02TDD143 pKa = 3.46YY144 pKa = 9.79TRR146 pKa = 11.84KK147 pKa = 9.53KK148 pKa = 10.49DD149 pKa = 3.43PVPRR153 pKa = 11.84VISPRR158 pKa = 11.84NPRR161 pKa = 11.84YY162 pKa = 8.55NVEE165 pKa = 3.58VGRR168 pKa = 11.84YY169 pKa = 8.5LRR171 pKa = 11.84TIEE174 pKa = 4.26EE175 pKa = 4.41PLFHH179 pKa = 7.06SLSQLFEE186 pKa = 4.11GKK188 pKa = 8.43RR189 pKa = 11.84TVFKK193 pKa = 10.91GMNAADD199 pKa = 4.42SGRR202 pKa = 11.84AMYY205 pKa = 10.08DD206 pKa = 2.66LWSSFRR212 pKa = 11.84KK213 pKa = 9.13PVAVGLDD220 pKa = 3.18ASRR223 pKa = 11.84FDD225 pKa = 3.56QHH227 pKa = 6.64VSKK230 pKa = 10.97CALQWEE236 pKa = 4.6HH237 pKa = 7.14AIYY240 pKa = 8.55PQCFTSSGDD249 pKa = 3.33RR250 pKa = 11.84AEE252 pKa = 4.61LRR254 pKa = 11.84RR255 pKa = 11.84LLKK258 pKa = 9.72WQVHH262 pKa = 5.09NKK264 pKa = 9.46CVGYY268 pKa = 10.33CADD271 pKa = 3.94GRR273 pKa = 11.84IKK275 pKa = 9.36YY276 pKa = 7.66TKK278 pKa = 9.85EE279 pKa = 3.56GTRR282 pKa = 11.84MSGDD286 pKa = 3.41MNTSLGNCVLMCSMIKK302 pKa = 10.05QYY304 pKa = 11.37SLDD307 pKa = 3.57RR308 pKa = 11.84GVRR311 pKa = 11.84TLLANNGDD319 pKa = 3.82DD320 pKa = 3.43CVVFMEE326 pKa = 4.63ADD328 pKa = 3.23EE329 pKa = 4.4LARR332 pKa = 11.84FSSGLDD338 pKa = 2.85EE339 pKa = 4.68WFRR342 pKa = 11.84AMGFNMVVEE351 pKa = 4.57PPCYY355 pKa = 9.21QFEE358 pKa = 4.42EE359 pKa = 4.53IEE361 pKa = 4.43FCQTHH366 pKa = 7.26PIYY369 pKa = 10.74VGPKK373 pKa = 8.16HH374 pKa = 6.94SDD376 pKa = 3.25YY377 pKa = 11.69LMVRR381 pKa = 11.84HH382 pKa = 6.27PKK384 pKa = 8.62WALAKK389 pKa = 10.66DD390 pKa = 3.77SMCIHH395 pKa = 7.12GFPTSKK401 pKa = 9.78IYY403 pKa = 10.36KK404 pKa = 8.74AWLDD408 pKa = 3.6AVGTGGLAMTGGVPIFQDD426 pKa = 5.37FYY428 pKa = 11.37STYY431 pKa = 9.9CKK433 pKa = 10.56YY434 pKa = 10.33GVRR437 pKa = 11.84GKK439 pKa = 7.3THH441 pKa = 6.07FHH443 pKa = 5.02EE444 pKa = 4.59QSWGVRR450 pKa = 11.84SLQQGMVRR458 pKa = 11.84KK459 pKa = 9.41YY460 pKa = 10.73GAVLPEE466 pKa = 4.21TRR468 pKa = 11.84ASFYY472 pKa = 10.04WAFGVLPDD480 pKa = 3.91EE481 pKa = 4.27QLAIEE486 pKa = 4.25DD487 pKa = 4.68FYY489 pKa = 11.64RR490 pKa = 11.84GVKK493 pKa = 9.97LDD495 pKa = 3.7EE496 pKa = 4.14AWRR499 pKa = 11.84DD500 pKa = 3.51EE501 pKa = 5.94LEE503 pKa = 4.25FQPLLPLL510 pKa = 4.0
Molecular weight: 58.55 kDa
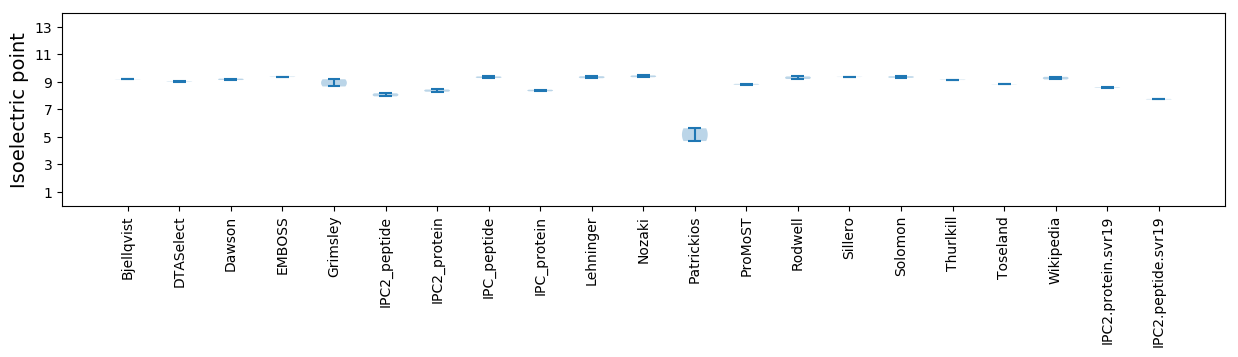
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGL0|A0A1L3KGL0_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 5 OX=1923292 PE=4 SV=1
MM1 pKa = 6.59VTIILEE7 pKa = 4.09AALISSVIVPLATTTALRR25 pKa = 11.84VSRR28 pKa = 11.84WMEE31 pKa = 3.52RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NALAKK40 pKa = 9.92EE41 pKa = 4.23CIARR45 pKa = 11.84FDD47 pKa = 3.9NLDD50 pKa = 3.38GSFANVLEE58 pKa = 4.47SARR61 pKa = 11.84QDD63 pKa = 3.31SARR66 pKa = 11.84RR67 pKa = 11.84DD68 pKa = 3.91AIEE71 pKa = 4.08ATAVTSAVLSNMGVVGVTVEE91 pKa = 4.19SACAQKK97 pKa = 10.49AAEE100 pKa = 4.23RR101 pKa = 11.84MRR103 pKa = 11.84EE104 pKa = 3.65QRR106 pKa = 11.84GWIRR110 pKa = 11.84IRR112 pKa = 11.84NHH114 pKa = 6.76RR115 pKa = 11.84LTEE118 pKa = 4.08LAFSAADD125 pKa = 3.32EE126 pKa = 4.86AYY128 pKa = 10.68NEE130 pKa = 4.08FGARR134 pKa = 11.84SLSKK138 pKa = 11.17ANDD141 pKa = 3.17LVTRR145 pKa = 11.84KK146 pKa = 9.4FLRR149 pKa = 11.84DD150 pKa = 3.53FFRR153 pKa = 11.84EE154 pKa = 4.25CKK156 pKa = 10.18DD157 pKa = 4.53LRR159 pKa = 11.84TKK161 pKa = 10.45DD162 pKa = 3.24ANRR165 pKa = 11.84AIEE168 pKa = 3.83IALPFSYY175 pKa = 10.38LPPAEE180 pKa = 4.28RR181 pKa = 11.84GSMQEE186 pKa = 4.86FVTLDD191 pKa = 3.45PFVEE195 pKa = 4.39RR196 pKa = 11.84VGPADD201 pKa = 4.86LRR203 pKa = 11.84VQQ205 pKa = 3.46
MM1 pKa = 6.59VTIILEE7 pKa = 4.09AALISSVIVPLATTTALRR25 pKa = 11.84VSRR28 pKa = 11.84WMEE31 pKa = 3.52RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NALAKK40 pKa = 9.92EE41 pKa = 4.23CIARR45 pKa = 11.84FDD47 pKa = 3.9NLDD50 pKa = 3.38GSFANVLEE58 pKa = 4.47SARR61 pKa = 11.84QDD63 pKa = 3.31SARR66 pKa = 11.84RR67 pKa = 11.84DD68 pKa = 3.91AIEE71 pKa = 4.08ATAVTSAVLSNMGVVGVTVEE91 pKa = 4.19SACAQKK97 pKa = 10.49AAEE100 pKa = 4.23RR101 pKa = 11.84MRR103 pKa = 11.84EE104 pKa = 3.65QRR106 pKa = 11.84GWIRR110 pKa = 11.84IRR112 pKa = 11.84NHH114 pKa = 6.76RR115 pKa = 11.84LTEE118 pKa = 4.08LAFSAADD125 pKa = 3.32EE126 pKa = 4.86AYY128 pKa = 10.68NEE130 pKa = 4.08FGARR134 pKa = 11.84SLSKK138 pKa = 11.17ANDD141 pKa = 3.17LVTRR145 pKa = 11.84KK146 pKa = 9.4FLRR149 pKa = 11.84DD150 pKa = 3.53FFRR153 pKa = 11.84EE154 pKa = 4.25CKK156 pKa = 10.18DD157 pKa = 4.53LRR159 pKa = 11.84TKK161 pKa = 10.45DD162 pKa = 3.24ANRR165 pKa = 11.84AIEE168 pKa = 3.83IALPFSYY175 pKa = 10.38LPPAEE180 pKa = 4.28RR181 pKa = 11.84GSMQEE186 pKa = 4.86FVTLDD191 pKa = 3.45PFVEE195 pKa = 4.39RR196 pKa = 11.84VGPADD201 pKa = 4.86LRR203 pKa = 11.84VQQ205 pKa = 3.46
Molecular weight: 22.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
715 |
205 |
510 |
357.5 |
40.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.811 ± 2.78 | 1.958 ± 0.258 |
5.734 ± 0.192 | 6.573 ± 0.642 |
5.455 ± 0.3 | 5.734 ± 1.208 |
1.958 ± 0.766 | 3.357 ± 0.793 |
5.734 ± 1.463 | 8.112 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.937 ± 0.259 | 2.797 ± 0.576 |
4.615 ± 0.88 | 3.217 ± 0.405 |
9.231 ± 1.544 | 6.434 ± 0.206 |
4.755 ± 0.318 | 7.972 ± 0.087 |
1.678 ± 0.366 | 2.937 ± 1.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
