
Thioflavicoccus mobilis 8321
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Thioflavicoccus; Thioflavicoccus mobilis
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
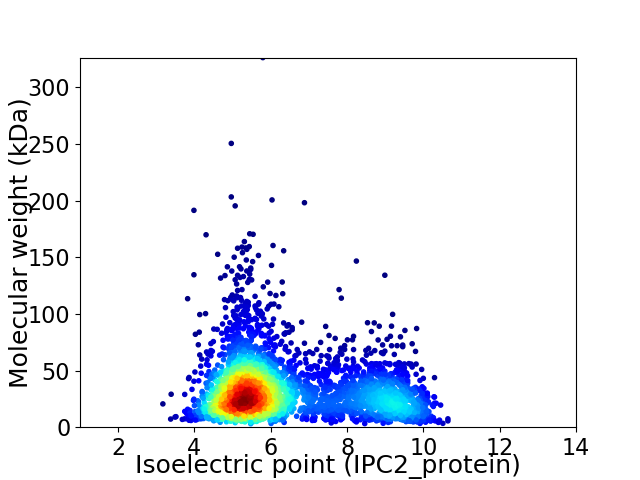
Virtual 2D-PAGE plot for 3528 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0GW24|L0GW24_9GAMM FeS assembly protein SufB OS=Thioflavicoccus mobilis 8321 OX=765912 GN=Thimo_2216 PE=3 SV=1
MM1 pKa = 7.43LPIILALCLFAGATQADD18 pKa = 3.88DD19 pKa = 5.06ALIQEE24 pKa = 4.74YY25 pKa = 10.49LSAPALPPASLTDD38 pKa = 3.46DD39 pKa = 3.63TVEE42 pKa = 4.12PEE44 pKa = 3.75VTIIEE49 pKa = 4.36RR50 pKa = 11.84GQDD53 pKa = 3.04VIYY56 pKa = 9.59EE57 pKa = 4.0YY58 pKa = 10.53RR59 pKa = 11.84VRR61 pKa = 11.84GQLYY65 pKa = 7.95MVKK68 pKa = 9.18VQPIAGPTYY77 pKa = 10.59YY78 pKa = 11.27LLDD81 pKa = 3.74TDD83 pKa = 5.35GDD85 pKa = 4.18GTLDD89 pKa = 3.58TRR91 pKa = 11.84DD92 pKa = 3.83DD93 pKa = 4.05RR94 pKa = 11.84PLNMATQQWLLFDD107 pKa = 3.89WW108 pKa = 4.61
MM1 pKa = 7.43LPIILALCLFAGATQADD18 pKa = 3.88DD19 pKa = 5.06ALIQEE24 pKa = 4.74YY25 pKa = 10.49LSAPALPPASLTDD38 pKa = 3.46DD39 pKa = 3.63TVEE42 pKa = 4.12PEE44 pKa = 3.75VTIIEE49 pKa = 4.36RR50 pKa = 11.84GQDD53 pKa = 3.04VIYY56 pKa = 9.59EE57 pKa = 4.0YY58 pKa = 10.53RR59 pKa = 11.84VRR61 pKa = 11.84GQLYY65 pKa = 7.95MVKK68 pKa = 9.18VQPIAGPTYY77 pKa = 10.59YY78 pKa = 11.27LLDD81 pKa = 3.74TDD83 pKa = 5.35GDD85 pKa = 4.18GTLDD89 pKa = 3.58TRR91 pKa = 11.84DD92 pKa = 3.83DD93 pKa = 4.05RR94 pKa = 11.84PLNMATQQWLLFDD107 pKa = 3.89WW108 pKa = 4.61
Molecular weight: 12.11 kDa
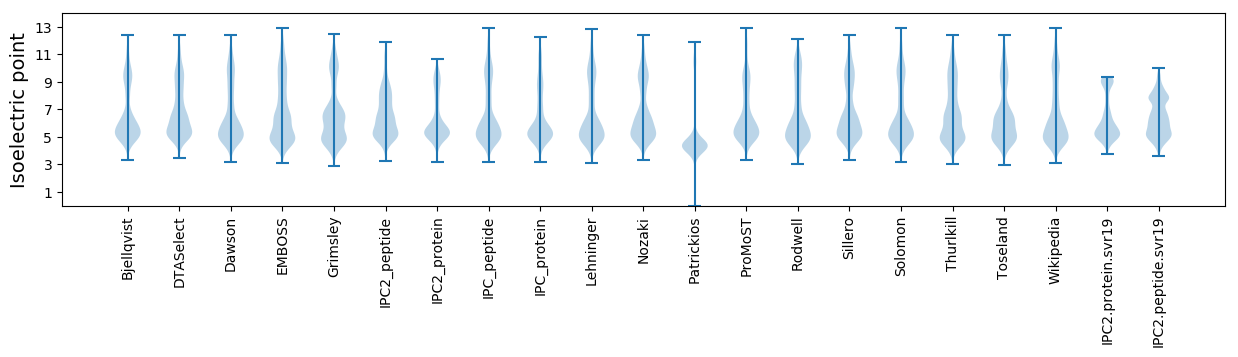
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0GT22|L0GT22_9GAMM Mg-chelatase subunit ChlD OS=Thioflavicoccus mobilis 8321 OX=765912 GN=Thimo_1115 PE=4 SV=1
MM1 pKa = 7.58SLPSSEE7 pKa = 4.11VRR9 pKa = 11.84SPSATAEE16 pKa = 4.06PPTADD21 pKa = 3.16RR22 pKa = 11.84SGRR25 pKa = 11.84AHH27 pKa = 5.93LRR29 pKa = 11.84EE30 pKa = 3.87RR31 pKa = 11.84AIRR34 pKa = 11.84WLIRR38 pKa = 11.84LIARR42 pKa = 11.84LPLAWIHH49 pKa = 6.45RR50 pKa = 11.84LGATLSQLIVWLPNRR65 pKa = 11.84QRR67 pKa = 11.84RR68 pKa = 11.84NALINIATCLPDD80 pKa = 4.25LSPTEE85 pKa = 4.17QVALRR90 pKa = 11.84NANLRR95 pKa = 11.84EE96 pKa = 3.86FGKK99 pKa = 9.04TYY101 pKa = 11.06LEE103 pKa = 5.0IGHH106 pKa = 7.1LWLRR110 pKa = 11.84PVDD113 pKa = 3.83EE114 pKa = 4.35VLGLIRR120 pKa = 11.84EE121 pKa = 4.27VRR123 pKa = 11.84GAEE126 pKa = 3.8LLRR129 pKa = 11.84RR130 pKa = 11.84HH131 pKa = 6.03GGKK134 pKa = 10.36GLIVLSPHH142 pKa = 5.56IGAWEE147 pKa = 3.66LAGLYY152 pKa = 10.48LSIQGPTAIFYY163 pKa = 10.47KK164 pKa = 8.66PQKK167 pKa = 10.32YY168 pKa = 10.43LDD170 pKa = 4.15DD171 pKa = 5.69LIVAARR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84GGARR183 pKa = 11.84LAPITAKK190 pKa = 10.6GIRR193 pKa = 11.84VLVQALEE200 pKa = 3.93RR201 pKa = 11.84GEE203 pKa = 4.07YY204 pKa = 10.42VGILPDD210 pKa = 4.57QEE212 pKa = 4.13PKK214 pKa = 9.25MDD216 pKa = 3.8KK217 pKa = 10.82GAVFAPFFGIPALTMLLVNRR237 pKa = 11.84LARR240 pKa = 11.84RR241 pKa = 11.84TGAPVIFMFAEE252 pKa = 4.27RR253 pKa = 11.84LPRR256 pKa = 11.84ARR258 pKa = 11.84GFRR261 pKa = 11.84IHH263 pKa = 7.07CLPAPDD269 pKa = 5.76GIDD272 pKa = 3.93SADD275 pKa = 3.6DD276 pKa = 3.67HH277 pKa = 6.15QAAAALNRR285 pKa = 11.84GIEE288 pKa = 4.02QCIGVCPAQFVWPYY302 pKa = 9.83KK303 pKa = 9.44RR304 pKa = 11.84FRR306 pKa = 11.84RR307 pKa = 11.84RR308 pKa = 11.84PEE310 pKa = 4.19GQPKK314 pKa = 10.53LYY316 pKa = 10.23TGPLDD321 pKa = 4.71DD322 pKa = 4.81AQTLARR328 pKa = 11.84LASDD332 pKa = 3.2QVGRR336 pKa = 11.84GTPTT340 pKa = 2.81
MM1 pKa = 7.58SLPSSEE7 pKa = 4.11VRR9 pKa = 11.84SPSATAEE16 pKa = 4.06PPTADD21 pKa = 3.16RR22 pKa = 11.84SGRR25 pKa = 11.84AHH27 pKa = 5.93LRR29 pKa = 11.84EE30 pKa = 3.87RR31 pKa = 11.84AIRR34 pKa = 11.84WLIRR38 pKa = 11.84LIARR42 pKa = 11.84LPLAWIHH49 pKa = 6.45RR50 pKa = 11.84LGATLSQLIVWLPNRR65 pKa = 11.84QRR67 pKa = 11.84RR68 pKa = 11.84NALINIATCLPDD80 pKa = 4.25LSPTEE85 pKa = 4.17QVALRR90 pKa = 11.84NANLRR95 pKa = 11.84EE96 pKa = 3.86FGKK99 pKa = 9.04TYY101 pKa = 11.06LEE103 pKa = 5.0IGHH106 pKa = 7.1LWLRR110 pKa = 11.84PVDD113 pKa = 3.83EE114 pKa = 4.35VLGLIRR120 pKa = 11.84EE121 pKa = 4.27VRR123 pKa = 11.84GAEE126 pKa = 3.8LLRR129 pKa = 11.84RR130 pKa = 11.84HH131 pKa = 6.03GGKK134 pKa = 10.36GLIVLSPHH142 pKa = 5.56IGAWEE147 pKa = 3.66LAGLYY152 pKa = 10.48LSIQGPTAIFYY163 pKa = 10.47KK164 pKa = 8.66PQKK167 pKa = 10.32YY168 pKa = 10.43LDD170 pKa = 4.15DD171 pKa = 5.69LIVAARR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84GGARR183 pKa = 11.84LAPITAKK190 pKa = 10.6GIRR193 pKa = 11.84VLVQALEE200 pKa = 3.93RR201 pKa = 11.84GEE203 pKa = 4.07YY204 pKa = 10.42VGILPDD210 pKa = 4.57QEE212 pKa = 4.13PKK214 pKa = 9.25MDD216 pKa = 3.8KK217 pKa = 10.82GAVFAPFFGIPALTMLLVNRR237 pKa = 11.84LARR240 pKa = 11.84RR241 pKa = 11.84TGAPVIFMFAEE252 pKa = 4.27RR253 pKa = 11.84LPRR256 pKa = 11.84ARR258 pKa = 11.84GFRR261 pKa = 11.84IHH263 pKa = 7.07CLPAPDD269 pKa = 5.76GIDD272 pKa = 3.93SADD275 pKa = 3.6DD276 pKa = 3.67HH277 pKa = 6.15QAAAALNRR285 pKa = 11.84GIEE288 pKa = 4.02QCIGVCPAQFVWPYY302 pKa = 9.83KK303 pKa = 9.44RR304 pKa = 11.84FRR306 pKa = 11.84RR307 pKa = 11.84RR308 pKa = 11.84PEE310 pKa = 4.19GQPKK314 pKa = 10.53LYY316 pKa = 10.23TGPLDD321 pKa = 4.71DD322 pKa = 4.81AQTLARR328 pKa = 11.84LASDD332 pKa = 3.2QVGRR336 pKa = 11.84GTPTT340 pKa = 2.81
Molecular weight: 37.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1133450 |
28 |
2948 |
321.3 |
35.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.049 ± 0.045 | 1.022 ± 0.014 |
5.891 ± 0.029 | 6.316 ± 0.042 |
3.335 ± 0.025 | 8.328 ± 0.036 |
2.239 ± 0.019 | 4.714 ± 0.029 |
2.467 ± 0.029 | 11.358 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.015 ± 0.018 | 2.237 ± 0.021 |
5.467 ± 0.033 | 3.43 ± 0.024 |
8.338 ± 0.046 | 4.804 ± 0.032 |
4.948 ± 0.031 | 7.222 ± 0.036 |
1.435 ± 0.019 | 2.386 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
