
Pistacia emaravirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Fimoviridae; Emaravirus; unclassified Emaravirus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
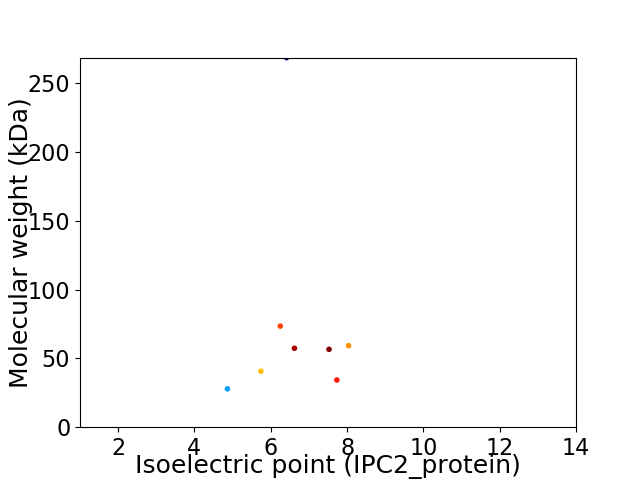
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A410JAQ1|A0A410JAQ1_9VIRU p6 protein OS=Pistacia emaravirus OX=2507689 PE=4 SV=1
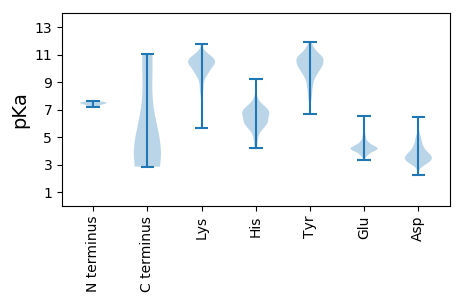
MM1 pKa = 7.49EE2 pKa = 5.01FKK4 pKa = 10.89SYY6 pKa = 10.77VRR8 pKa = 11.84TFNACRR14 pKa = 11.84KK15 pKa = 9.76GNVSDD20 pKa = 4.26YY21 pKa = 10.98YY22 pKa = 10.56IEE24 pKa = 5.06FDD26 pKa = 3.29HH27 pKa = 6.93QLFFSIVEE35 pKa = 4.07RR36 pKa = 11.84MDD38 pKa = 3.62TEE40 pKa = 4.11THH42 pKa = 5.97DD43 pKa = 3.06QFLRR47 pKa = 11.84FGEE50 pKa = 4.75LYY52 pKa = 10.65DD53 pKa = 5.43DD54 pKa = 4.5GNIDD58 pKa = 3.13IPEE61 pKa = 4.13IGRR64 pKa = 11.84TVNEE68 pKa = 4.26IVLDD72 pKa = 4.77DD73 pKa = 3.41ITIKK77 pKa = 10.35KK78 pKa = 9.99KK79 pKa = 10.18AMRR82 pKa = 11.84YY83 pKa = 7.34HH84 pKa = 6.22FRR86 pKa = 11.84YY87 pKa = 9.46IQILHH92 pKa = 6.77SILGIVDD99 pKa = 4.08AFNDD103 pKa = 3.44EE104 pKa = 4.3MSPDD108 pKa = 3.14FHH110 pKa = 9.21IFGTLVLKK118 pKa = 11.01NKK120 pKa = 9.77LLPFIPDD127 pKa = 3.25NSDD130 pKa = 3.49CLLLSFDD137 pKa = 4.93IIKK140 pKa = 10.59NLIWEE145 pKa = 4.23IVIGYY150 pKa = 9.72DD151 pKa = 4.03PIDD154 pKa = 3.95IIEE157 pKa = 4.56KK158 pKa = 9.63YY159 pKa = 10.19KK160 pKa = 10.75SRR162 pKa = 11.84QLKK165 pKa = 9.47SVIYY169 pKa = 10.88NEE171 pKa = 4.27DD172 pKa = 3.06TVAYY176 pKa = 8.84QFQVLAISYY185 pKa = 8.42LAIRR189 pKa = 11.84ASSMVDD195 pKa = 3.02KK196 pKa = 10.63EE197 pKa = 3.92KK198 pKa = 11.31SKK200 pKa = 11.24NKK202 pKa = 10.29SYY204 pKa = 11.38NDD206 pKa = 3.56SNQVTIEE213 pKa = 4.46DD214 pKa = 4.03GNKK217 pKa = 10.05LSEE220 pKa = 4.67LEE222 pKa = 4.19SDD224 pKa = 3.95SKK226 pKa = 11.5SIASSSHH233 pKa = 5.91QDD235 pKa = 3.35DD236 pKa = 4.76LMSKK240 pKa = 10.39
MM1 pKa = 7.49EE2 pKa = 5.01FKK4 pKa = 10.89SYY6 pKa = 10.77VRR8 pKa = 11.84TFNACRR14 pKa = 11.84KK15 pKa = 9.76GNVSDD20 pKa = 4.26YY21 pKa = 10.98YY22 pKa = 10.56IEE24 pKa = 5.06FDD26 pKa = 3.29HH27 pKa = 6.93QLFFSIVEE35 pKa = 4.07RR36 pKa = 11.84MDD38 pKa = 3.62TEE40 pKa = 4.11THH42 pKa = 5.97DD43 pKa = 3.06QFLRR47 pKa = 11.84FGEE50 pKa = 4.75LYY52 pKa = 10.65DD53 pKa = 5.43DD54 pKa = 4.5GNIDD58 pKa = 3.13IPEE61 pKa = 4.13IGRR64 pKa = 11.84TVNEE68 pKa = 4.26IVLDD72 pKa = 4.77DD73 pKa = 3.41ITIKK77 pKa = 10.35KK78 pKa = 9.99KK79 pKa = 10.18AMRR82 pKa = 11.84YY83 pKa = 7.34HH84 pKa = 6.22FRR86 pKa = 11.84YY87 pKa = 9.46IQILHH92 pKa = 6.77SILGIVDD99 pKa = 4.08AFNDD103 pKa = 3.44EE104 pKa = 4.3MSPDD108 pKa = 3.14FHH110 pKa = 9.21IFGTLVLKK118 pKa = 11.01NKK120 pKa = 9.77LLPFIPDD127 pKa = 3.25NSDD130 pKa = 3.49CLLLSFDD137 pKa = 4.93IIKK140 pKa = 10.59NLIWEE145 pKa = 4.23IVIGYY150 pKa = 9.72DD151 pKa = 4.03PIDD154 pKa = 3.95IIEE157 pKa = 4.56KK158 pKa = 9.63YY159 pKa = 10.19KK160 pKa = 10.75SRR162 pKa = 11.84QLKK165 pKa = 9.47SVIYY169 pKa = 10.88NEE171 pKa = 4.27DD172 pKa = 3.06TVAYY176 pKa = 8.84QFQVLAISYY185 pKa = 8.42LAIRR189 pKa = 11.84ASSMVDD195 pKa = 3.02KK196 pKa = 10.63EE197 pKa = 3.92KK198 pKa = 11.31SKK200 pKa = 11.24NKK202 pKa = 10.29SYY204 pKa = 11.38NDD206 pKa = 3.56SNQVTIEE213 pKa = 4.46DD214 pKa = 4.03GNKK217 pKa = 10.05LSEE220 pKa = 4.67LEE222 pKa = 4.19SDD224 pKa = 3.95SKK226 pKa = 11.5SIASSSHH233 pKa = 5.91QDD235 pKa = 3.35DD236 pKa = 4.76LMSKK240 pKa = 10.39
Molecular weight: 27.89 kDa
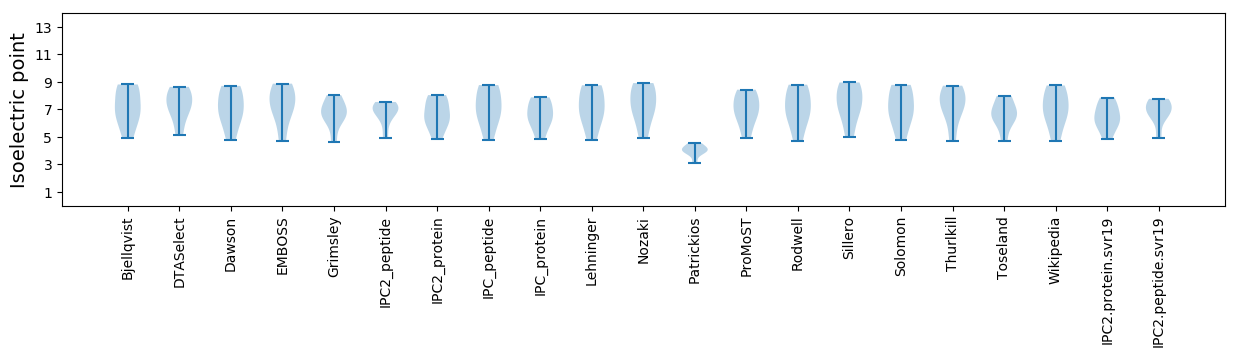
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A410JAN1|A0A410JAN1_9VIRU p5 variant B OS=Pistacia emaravirus OX=2507689 PE=4 SV=1
MM1 pKa = 7.49IHH3 pKa = 5.9NQIIKK8 pKa = 9.85KK9 pKa = 7.87QRR11 pKa = 11.84KK12 pKa = 7.22KK13 pKa = 10.8QLVSYY18 pKa = 9.43LAKK21 pKa = 9.74NANMEE26 pKa = 4.45HH27 pKa = 6.6IKK29 pKa = 10.43PYY31 pKa = 8.71TNSIGGCTIKK41 pKa = 10.72LSNKK45 pKa = 9.49EE46 pKa = 4.24KK47 pKa = 10.31ISDD50 pKa = 3.8YY51 pKa = 10.95KK52 pKa = 11.02GLSSFEE58 pKa = 4.05EE59 pKa = 4.17NIYY62 pKa = 10.72NQIKK66 pKa = 10.8NKK68 pKa = 9.95FNNCEE73 pKa = 4.1CFEE76 pKa = 4.74KK77 pKa = 10.89DD78 pKa = 3.47VFFDD82 pKa = 4.42EE83 pKa = 5.04DD84 pKa = 3.63SKK86 pKa = 11.03MPKK89 pKa = 9.36NASISYY95 pKa = 10.25IEE97 pKa = 5.18INDD100 pKa = 3.28QFPRR104 pKa = 11.84YY105 pKa = 8.62MYY107 pKa = 9.15TIKK110 pKa = 10.75KK111 pKa = 8.38MRR113 pKa = 11.84SKK115 pKa = 10.96YY116 pKa = 10.2KK117 pKa = 10.68YY118 pKa = 10.01SLTNIDD124 pKa = 5.17DD125 pKa = 4.36LLFSIIKK132 pKa = 9.51NHH134 pKa = 5.3LHH136 pKa = 5.61NFIIRR141 pKa = 11.84EE142 pKa = 4.01FEE144 pKa = 3.89SDD146 pKa = 3.04INEE149 pKa = 3.57INIIIKK155 pKa = 10.42GFLHH159 pKa = 6.54IPKK162 pKa = 9.4QFVMVKK168 pKa = 10.04INNKK172 pKa = 9.37NLLINDD178 pKa = 4.1TKK180 pKa = 11.43YY181 pKa = 10.07NWEE184 pKa = 4.39NIISSTTLCDD194 pKa = 3.34LRR196 pKa = 11.84SILSHH201 pKa = 6.77FDD203 pKa = 3.36DD204 pKa = 4.7MRR206 pKa = 11.84DD207 pKa = 3.62FYY209 pKa = 11.11YY210 pKa = 10.88KK211 pKa = 10.23KK212 pKa = 10.46VQGTNIVKK220 pKa = 10.32SIYY223 pKa = 10.91SFINDD228 pKa = 3.14NKK230 pKa = 10.92SLINHH235 pKa = 6.16VLPSIKK241 pKa = 9.69TSFKK245 pKa = 10.64TYY247 pKa = 10.46DD248 pKa = 3.61DD249 pKa = 4.44CVLKK253 pKa = 10.76YY254 pKa = 10.58INAHH258 pKa = 5.56LKK260 pKa = 9.61KK261 pKa = 10.49AYY263 pKa = 10.17NEE265 pKa = 4.38SNYY268 pKa = 11.04SLVFDD273 pKa = 4.3SSTIQEE279 pKa = 4.19FLIEE283 pKa = 4.37TEE285 pKa = 4.2SCSDD289 pKa = 3.58FSLEE293 pKa = 3.7VRR295 pKa = 11.84KK296 pKa = 9.98AIHH299 pKa = 6.69EE300 pKa = 4.35DD301 pKa = 3.14VTKK304 pKa = 10.54QLEE307 pKa = 4.21GLKK310 pKa = 8.49TQFSEE315 pKa = 4.11YY316 pKa = 10.42DD317 pKa = 3.4ILKK320 pKa = 8.92MPCLEE325 pKa = 4.6LVNQRR330 pKa = 11.84LVPIKK335 pKa = 10.52HH336 pKa = 6.31NKK338 pKa = 8.91NKK340 pKa = 10.48LNVSGSCEE348 pKa = 3.71QSVAFPLNIDD358 pKa = 4.52EE359 pKa = 4.35IVKK362 pKa = 9.98YY363 pKa = 10.11PPGEE367 pKa = 4.02NPIQDD372 pKa = 3.38NFYY375 pKa = 10.75YY376 pKa = 10.75GLPNTLEE383 pKa = 4.09ISTQYY388 pKa = 10.98LNSAMRR394 pKa = 11.84KK395 pKa = 9.36IIQEE399 pKa = 4.11EE400 pKa = 4.12KK401 pKa = 10.73DD402 pKa = 3.36IDD404 pKa = 4.92DD405 pKa = 3.56IDD407 pKa = 4.64YY408 pKa = 10.83IAGYY412 pKa = 8.27PFKK415 pKa = 10.43RR416 pKa = 11.84SYY418 pKa = 10.7KK419 pKa = 9.75ILIFSILKK427 pKa = 8.3KK428 pKa = 9.86QNYY431 pKa = 9.07NIRR434 pKa = 11.84ISHH437 pKa = 6.93LFLHH441 pKa = 5.91YY442 pKa = 10.19FCYY445 pKa = 10.56YY446 pKa = 9.47MYY448 pKa = 10.05AASVASIRR456 pKa = 11.84VGVDD460 pKa = 2.86MIYY463 pKa = 10.23EE464 pKa = 3.98QKK466 pKa = 10.82QIDD469 pKa = 3.75RR470 pKa = 11.84ARR472 pKa = 11.84EE473 pKa = 3.69FSKK476 pKa = 11.16KK477 pKa = 10.04NISVIRR483 pKa = 11.84NLVQSHH489 pKa = 5.55KK490 pKa = 10.53HH491 pKa = 5.96LFLQNQCVSITLAA504 pKa = 3.73
MM1 pKa = 7.49IHH3 pKa = 5.9NQIIKK8 pKa = 9.85KK9 pKa = 7.87QRR11 pKa = 11.84KK12 pKa = 7.22KK13 pKa = 10.8QLVSYY18 pKa = 9.43LAKK21 pKa = 9.74NANMEE26 pKa = 4.45HH27 pKa = 6.6IKK29 pKa = 10.43PYY31 pKa = 8.71TNSIGGCTIKK41 pKa = 10.72LSNKK45 pKa = 9.49EE46 pKa = 4.24KK47 pKa = 10.31ISDD50 pKa = 3.8YY51 pKa = 10.95KK52 pKa = 11.02GLSSFEE58 pKa = 4.05EE59 pKa = 4.17NIYY62 pKa = 10.72NQIKK66 pKa = 10.8NKK68 pKa = 9.95FNNCEE73 pKa = 4.1CFEE76 pKa = 4.74KK77 pKa = 10.89DD78 pKa = 3.47VFFDD82 pKa = 4.42EE83 pKa = 5.04DD84 pKa = 3.63SKK86 pKa = 11.03MPKK89 pKa = 9.36NASISYY95 pKa = 10.25IEE97 pKa = 5.18INDD100 pKa = 3.28QFPRR104 pKa = 11.84YY105 pKa = 8.62MYY107 pKa = 9.15TIKK110 pKa = 10.75KK111 pKa = 8.38MRR113 pKa = 11.84SKK115 pKa = 10.96YY116 pKa = 10.2KK117 pKa = 10.68YY118 pKa = 10.01SLTNIDD124 pKa = 5.17DD125 pKa = 4.36LLFSIIKK132 pKa = 9.51NHH134 pKa = 5.3LHH136 pKa = 5.61NFIIRR141 pKa = 11.84EE142 pKa = 4.01FEE144 pKa = 3.89SDD146 pKa = 3.04INEE149 pKa = 3.57INIIIKK155 pKa = 10.42GFLHH159 pKa = 6.54IPKK162 pKa = 9.4QFVMVKK168 pKa = 10.04INNKK172 pKa = 9.37NLLINDD178 pKa = 4.1TKK180 pKa = 11.43YY181 pKa = 10.07NWEE184 pKa = 4.39NIISSTTLCDD194 pKa = 3.34LRR196 pKa = 11.84SILSHH201 pKa = 6.77FDD203 pKa = 3.36DD204 pKa = 4.7MRR206 pKa = 11.84DD207 pKa = 3.62FYY209 pKa = 11.11YY210 pKa = 10.88KK211 pKa = 10.23KK212 pKa = 10.46VQGTNIVKK220 pKa = 10.32SIYY223 pKa = 10.91SFINDD228 pKa = 3.14NKK230 pKa = 10.92SLINHH235 pKa = 6.16VLPSIKK241 pKa = 9.69TSFKK245 pKa = 10.64TYY247 pKa = 10.46DD248 pKa = 3.61DD249 pKa = 4.44CVLKK253 pKa = 10.76YY254 pKa = 10.58INAHH258 pKa = 5.56LKK260 pKa = 9.61KK261 pKa = 10.49AYY263 pKa = 10.17NEE265 pKa = 4.38SNYY268 pKa = 11.04SLVFDD273 pKa = 4.3SSTIQEE279 pKa = 4.19FLIEE283 pKa = 4.37TEE285 pKa = 4.2SCSDD289 pKa = 3.58FSLEE293 pKa = 3.7VRR295 pKa = 11.84KK296 pKa = 9.98AIHH299 pKa = 6.69EE300 pKa = 4.35DD301 pKa = 3.14VTKK304 pKa = 10.54QLEE307 pKa = 4.21GLKK310 pKa = 8.49TQFSEE315 pKa = 4.11YY316 pKa = 10.42DD317 pKa = 3.4ILKK320 pKa = 8.92MPCLEE325 pKa = 4.6LVNQRR330 pKa = 11.84LVPIKK335 pKa = 10.52HH336 pKa = 6.31NKK338 pKa = 8.91NKK340 pKa = 10.48LNVSGSCEE348 pKa = 3.71QSVAFPLNIDD358 pKa = 4.52EE359 pKa = 4.35IVKK362 pKa = 9.98YY363 pKa = 10.11PPGEE367 pKa = 4.02NPIQDD372 pKa = 3.38NFYY375 pKa = 10.75YY376 pKa = 10.75GLPNTLEE383 pKa = 4.09ISTQYY388 pKa = 10.98LNSAMRR394 pKa = 11.84KK395 pKa = 9.36IIQEE399 pKa = 4.11EE400 pKa = 4.12KK401 pKa = 10.73DD402 pKa = 3.36IDD404 pKa = 4.92DD405 pKa = 3.56IDD407 pKa = 4.64YY408 pKa = 10.83IAGYY412 pKa = 8.27PFKK415 pKa = 10.43RR416 pKa = 11.84SYY418 pKa = 10.7KK419 pKa = 9.75ILIFSILKK427 pKa = 8.3KK428 pKa = 9.86QNYY431 pKa = 9.07NIRR434 pKa = 11.84ISHH437 pKa = 6.93LFLHH441 pKa = 5.91YY442 pKa = 10.19FCYY445 pKa = 10.56YY446 pKa = 9.47MYY448 pKa = 10.05AASVASIRR456 pKa = 11.84VGVDD460 pKa = 2.86MIYY463 pKa = 10.23EE464 pKa = 3.98QKK466 pKa = 10.82QIDD469 pKa = 3.75RR470 pKa = 11.84ARR472 pKa = 11.84EE473 pKa = 3.69FSKK476 pKa = 11.16KK477 pKa = 10.04NISVIRR483 pKa = 11.84NLVQSHH489 pKa = 5.55KK490 pKa = 10.53HH491 pKa = 5.96LFLQNQCVSITLAA504 pKa = 3.73
Molecular weight: 59.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5314 |
240 |
2299 |
664.3 |
77.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.632 ± 0.558 | 1.844 ± 0.37 |
6.361 ± 0.388 | 6.022 ± 0.481 |
4.686 ± 0.307 | 3.049 ± 0.44 |
2.465 ± 0.208 | 10.181 ± 0.502 |
8.863 ± 0.286 | 8.863 ± 0.554 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.011 ± 0.374 | 7.49 ± 0.396 |
3.105 ± 0.164 | 2.616 ± 0.256 |
3.406 ± 0.188 | 7.697 ± 0.42 |
5.702 ± 0.496 | 4.535 ± 0.457 |
0.452 ± 0.149 | 6.022 ± 0.515 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
