
Dishui Lake virophage 2
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
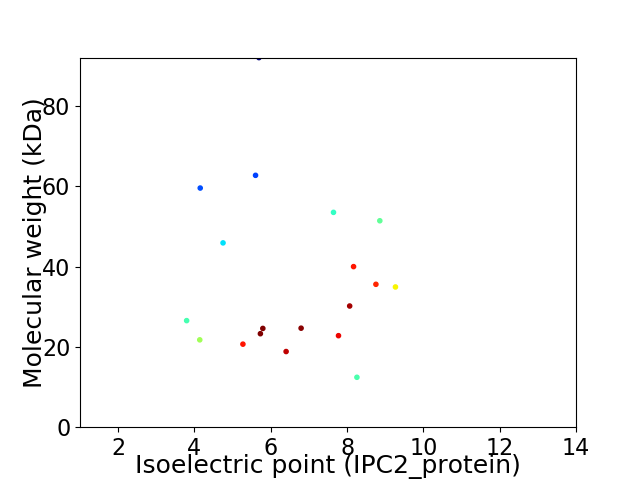
Virtual 2D-PAGE plot for 19 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6XN33|A0A6G6XN33_9VIRU Uncharacterized protein OS=Dishui Lake virophage 2 OX=2704064 PE=4 SV=1
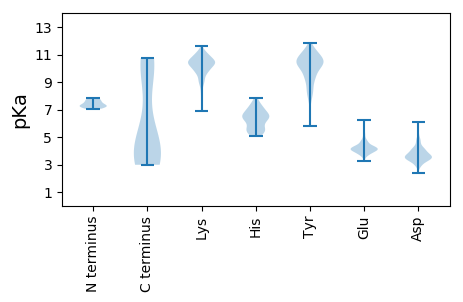
MM1 pKa = 7.26ATNSSFGYY9 pKa = 10.07LPVGSIIYY17 pKa = 9.5YY18 pKa = 10.01AGNGEE23 pKa = 4.06IPSTFLRR30 pKa = 11.84CDD32 pKa = 2.96GTNYY36 pKa = 10.23DD37 pKa = 3.77RR38 pKa = 11.84EE39 pKa = 4.56TYY41 pKa = 10.56SVLFDD46 pKa = 3.9VLGTSYY52 pKa = 11.53GSTSATNFKK61 pKa = 10.92VPDD64 pKa = 4.92LINYY68 pKa = 8.01YY69 pKa = 8.98YY70 pKa = 10.17IKK72 pKa = 10.8GGAQSSGVPSQAVIDD87 pKa = 4.1VPPFLLNSATLPSLAQGNFVFDD109 pKa = 3.78SWNLTANINNGVWFSNSTPSVDD131 pKa = 3.73VVPSGSDD138 pKa = 3.17DD139 pKa = 3.63TVKK142 pKa = 11.03ANSTDD147 pKa = 3.37QNNYY151 pKa = 7.94TGAVTGGTIGFTNPAQEE168 pKa = 4.77TIDD171 pKa = 4.19LAPDD175 pKa = 3.39PTTTVEE181 pKa = 4.17LQALSLIPLIKK192 pKa = 10.02AWYY195 pKa = 9.3DD196 pKa = 3.55FLPPTYY202 pKa = 9.81TPLPVSTDD210 pKa = 2.71SGIRR214 pKa = 11.84VDD216 pKa = 4.78KK217 pKa = 11.32VNEE220 pKa = 4.05TEE222 pKa = 4.27YY223 pKa = 8.58TTNPTGIYY231 pKa = 10.13LDD233 pKa = 4.81DD234 pKa = 4.53PQLSGFVFEE243 pKa = 5.65KK244 pKa = 9.76PTFF247 pKa = 3.68
MM1 pKa = 7.26ATNSSFGYY9 pKa = 10.07LPVGSIIYY17 pKa = 9.5YY18 pKa = 10.01AGNGEE23 pKa = 4.06IPSTFLRR30 pKa = 11.84CDD32 pKa = 2.96GTNYY36 pKa = 10.23DD37 pKa = 3.77RR38 pKa = 11.84EE39 pKa = 4.56TYY41 pKa = 10.56SVLFDD46 pKa = 3.9VLGTSYY52 pKa = 11.53GSTSATNFKK61 pKa = 10.92VPDD64 pKa = 4.92LINYY68 pKa = 8.01YY69 pKa = 8.98YY70 pKa = 10.17IKK72 pKa = 10.8GGAQSSGVPSQAVIDD87 pKa = 4.1VPPFLLNSATLPSLAQGNFVFDD109 pKa = 3.78SWNLTANINNGVWFSNSTPSVDD131 pKa = 3.73VVPSGSDD138 pKa = 3.17DD139 pKa = 3.63TVKK142 pKa = 11.03ANSTDD147 pKa = 3.37QNNYY151 pKa = 7.94TGAVTGGTIGFTNPAQEE168 pKa = 4.77TIDD171 pKa = 4.19LAPDD175 pKa = 3.39PTTTVEE181 pKa = 4.17LQALSLIPLIKK192 pKa = 10.02AWYY195 pKa = 9.3DD196 pKa = 3.55FLPPTYY202 pKa = 9.81TPLPVSTDD210 pKa = 2.71SGIRR214 pKa = 11.84VDD216 pKa = 4.78KK217 pKa = 11.32VNEE220 pKa = 4.05TEE222 pKa = 4.27YY223 pKa = 8.58TTNPTGIYY231 pKa = 10.13LDD233 pKa = 4.81DD234 pKa = 4.53PQLSGFVFEE243 pKa = 5.65KK244 pKa = 9.76PTFF247 pKa = 3.68
Molecular weight: 26.56 kDa
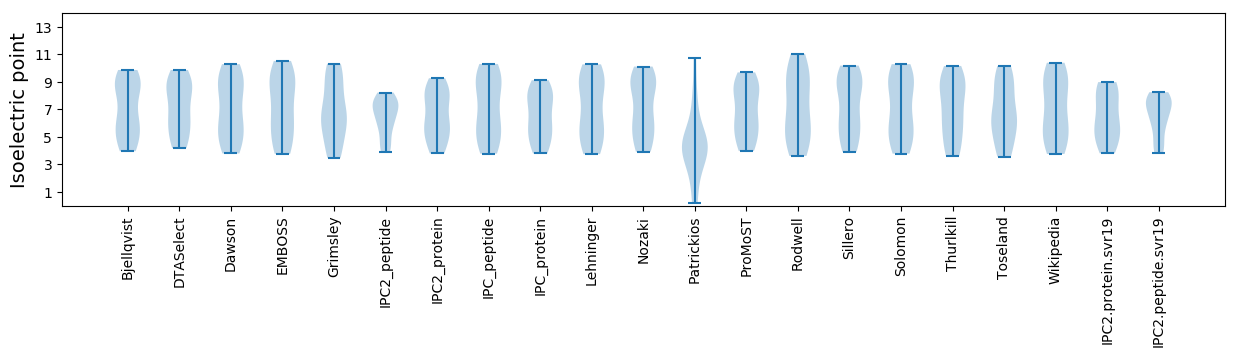
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6XME2|A0A6G6XME2_9VIRU Collar domain-containing protein OS=Dishui Lake virophage 2 OX=2704064 PE=4 SV=1
MM1 pKa = 7.12PQANITYY8 pKa = 8.74DD9 pKa = 2.89IPYY12 pKa = 8.33NRR14 pKa = 11.84KK15 pKa = 8.2MVDD18 pKa = 2.73ILRR21 pKa = 11.84EE22 pKa = 3.79MDD24 pKa = 3.21EE25 pKa = 4.2KK26 pKa = 10.69HH27 pKa = 5.12WRR29 pKa = 11.84KK30 pKa = 10.32AGDD33 pKa = 3.31AYY35 pKa = 10.73APTMFSEE42 pKa = 4.47KK43 pKa = 10.44LGNFHH48 pKa = 6.62GAKK51 pKa = 9.8IGGGSPANQQYY62 pKa = 9.62IHH64 pKa = 6.81SGNSPAYY71 pKa = 9.23PPVNMNSGMAVSSGGMYY88 pKa = 10.21SGIDD92 pKa = 3.37GAVGGKK98 pKa = 10.15YY99 pKa = 10.45SVDD102 pKa = 3.13KK103 pKa = 10.98FVGDD107 pKa = 3.96FKK109 pKa = 11.43KK110 pKa = 10.68VGKK113 pKa = 9.58LLKK116 pKa = 10.12PVAKK120 pKa = 10.07PILTALTNKK129 pKa = 9.8AVAKK133 pKa = 10.5LGGFSEE139 pKa = 5.13LMTGCGRR146 pKa = 11.84PAPKK150 pKa = 10.67NKK152 pKa = 9.28MDD154 pKa = 3.57VVDD157 pKa = 4.32FLKK160 pKa = 10.88PLGAKK165 pKa = 9.74KK166 pKa = 10.08SHH168 pKa = 5.85SLNRR172 pKa = 11.84LHH174 pKa = 7.0EE175 pKa = 4.22LVAPYY180 pKa = 10.55RR181 pKa = 11.84MEE183 pKa = 4.21GGKK186 pKa = 10.62YY187 pKa = 10.27SVDD190 pKa = 2.79KK191 pKa = 10.41FVRR194 pKa = 11.84DD195 pKa = 3.92FGKK198 pKa = 9.92IGKK201 pKa = 9.07LIKK204 pKa = 10.13PVAKK208 pKa = 10.06PILKK212 pKa = 10.34ALTDD216 pKa = 3.6KK217 pKa = 11.22AVAKK221 pKa = 9.23VTGAGMDD228 pKa = 3.58EE229 pKa = 4.6EE230 pKa = 5.22IEE232 pKa = 4.14LVRR235 pKa = 11.84PMEE238 pKa = 4.03MAEE241 pKa = 4.15TKK243 pKa = 10.83GKK245 pKa = 9.91GKK247 pKa = 10.55KK248 pKa = 8.87GGKK251 pKa = 9.22YY252 pKa = 10.42SVDD255 pKa = 3.15KK256 pKa = 10.6FVKK259 pKa = 10.49DD260 pKa = 3.77FGKK263 pKa = 10.03IGKK266 pKa = 9.07LIKK269 pKa = 10.07PVAKK273 pKa = 10.01PIVKK277 pKa = 10.27ALTDD281 pKa = 3.54KK282 pKa = 11.18AVAKK286 pKa = 10.36IGGGGGRR293 pKa = 11.84AKK295 pKa = 10.32RR296 pKa = 11.84AEE298 pKa = 3.98IVKK301 pKa = 10.36KK302 pKa = 10.86VMAEE306 pKa = 3.71KK307 pKa = 10.2GMKK310 pKa = 9.32MIEE313 pKa = 3.49ASKK316 pKa = 9.99YY317 pKa = 8.27VKK319 pKa = 10.34EE320 pKa = 4.26HH321 pKa = 5.73GLYY324 pKa = 10.57
MM1 pKa = 7.12PQANITYY8 pKa = 8.74DD9 pKa = 2.89IPYY12 pKa = 8.33NRR14 pKa = 11.84KK15 pKa = 8.2MVDD18 pKa = 2.73ILRR21 pKa = 11.84EE22 pKa = 3.79MDD24 pKa = 3.21EE25 pKa = 4.2KK26 pKa = 10.69HH27 pKa = 5.12WRR29 pKa = 11.84KK30 pKa = 10.32AGDD33 pKa = 3.31AYY35 pKa = 10.73APTMFSEE42 pKa = 4.47KK43 pKa = 10.44LGNFHH48 pKa = 6.62GAKK51 pKa = 9.8IGGGSPANQQYY62 pKa = 9.62IHH64 pKa = 6.81SGNSPAYY71 pKa = 9.23PPVNMNSGMAVSSGGMYY88 pKa = 10.21SGIDD92 pKa = 3.37GAVGGKK98 pKa = 10.15YY99 pKa = 10.45SVDD102 pKa = 3.13KK103 pKa = 10.98FVGDD107 pKa = 3.96FKK109 pKa = 11.43KK110 pKa = 10.68VGKK113 pKa = 9.58LLKK116 pKa = 10.12PVAKK120 pKa = 10.07PILTALTNKK129 pKa = 9.8AVAKK133 pKa = 10.5LGGFSEE139 pKa = 5.13LMTGCGRR146 pKa = 11.84PAPKK150 pKa = 10.67NKK152 pKa = 9.28MDD154 pKa = 3.57VVDD157 pKa = 4.32FLKK160 pKa = 10.88PLGAKK165 pKa = 9.74KK166 pKa = 10.08SHH168 pKa = 5.85SLNRR172 pKa = 11.84LHH174 pKa = 7.0EE175 pKa = 4.22LVAPYY180 pKa = 10.55RR181 pKa = 11.84MEE183 pKa = 4.21GGKK186 pKa = 10.62YY187 pKa = 10.27SVDD190 pKa = 2.79KK191 pKa = 10.41FVRR194 pKa = 11.84DD195 pKa = 3.92FGKK198 pKa = 9.92IGKK201 pKa = 9.07LIKK204 pKa = 10.13PVAKK208 pKa = 10.06PILKK212 pKa = 10.34ALTDD216 pKa = 3.6KK217 pKa = 11.22AVAKK221 pKa = 9.23VTGAGMDD228 pKa = 3.58EE229 pKa = 4.6EE230 pKa = 5.22IEE232 pKa = 4.14LVRR235 pKa = 11.84PMEE238 pKa = 4.03MAEE241 pKa = 4.15TKK243 pKa = 10.83GKK245 pKa = 9.91GKK247 pKa = 10.55KK248 pKa = 8.87GGKK251 pKa = 9.22YY252 pKa = 10.42SVDD255 pKa = 3.15KK256 pKa = 10.6FVKK259 pKa = 10.49DD260 pKa = 3.77FGKK263 pKa = 10.03IGKK266 pKa = 9.07LIKK269 pKa = 10.07PVAKK273 pKa = 10.01PIVKK277 pKa = 10.27ALTDD281 pKa = 3.54KK282 pKa = 11.18AVAKK286 pKa = 10.36IGGGGGRR293 pKa = 11.84AKK295 pKa = 10.32RR296 pKa = 11.84AEE298 pKa = 3.98IVKK301 pKa = 10.36KK302 pKa = 10.86VMAEE306 pKa = 3.71KK307 pKa = 10.2GMKK310 pKa = 9.32MIEE313 pKa = 3.49ASKK316 pKa = 9.99YY317 pKa = 8.27VKK319 pKa = 10.34EE320 pKa = 4.26HH321 pKa = 5.73GLYY324 pKa = 10.57
Molecular weight: 34.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6250 |
107 |
792 |
328.9 |
36.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.208 ± 0.472 | 1.088 ± 0.258 |
5.808 ± 0.35 | 5.936 ± 0.895 |
4.416 ± 0.378 | 6.608 ± 0.619 |
1.152 ± 0.213 | 6.496 ± 0.383 |
8.368 ± 1.286 | 8.416 ± 0.437 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.323 | 6.304 ± 0.598 |
4.624 ± 0.485 | 3.6 ± 0.373 |
3.616 ± 0.397 | 7.024 ± 0.766 |
6.848 ± 0.651 | 5.728 ± 0.362 |
1.232 ± 0.21 | 4.368 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
