
Turnip rosette virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
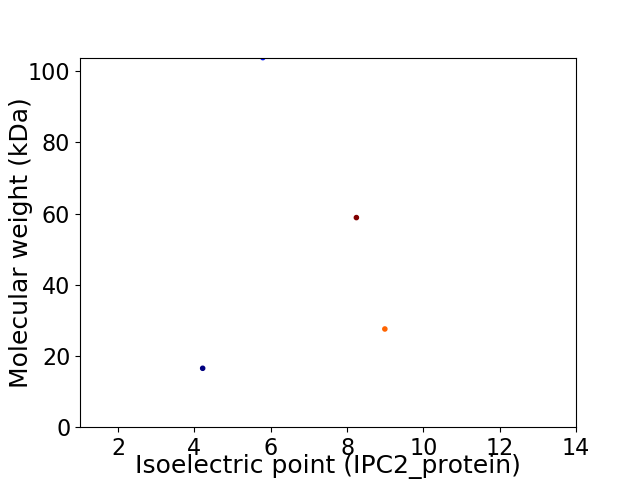
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4Z703|S4Z703_9VIRU N-terminal protein OS=Turnip rosette virus OX=218923 PE=4 SV=1
MM1 pKa = 7.2SRR3 pKa = 11.84VATIEE8 pKa = 3.82IYY10 pKa = 10.63NEE12 pKa = 3.42NGIIVARR19 pKa = 11.84KK20 pKa = 7.5KK21 pKa = 9.37TSGPHH26 pKa = 6.22ALLEE30 pKa = 4.23LFNGKK35 pKa = 8.18QKK37 pKa = 10.76YY38 pKa = 9.55DD39 pKa = 3.44QVSEE43 pKa = 4.45LFVIWVCEE51 pKa = 4.03EE52 pKa = 4.0CGKK55 pKa = 8.43TVYY58 pKa = 8.96STCEE62 pKa = 3.36FKK64 pKa = 11.22GIVFVRR70 pKa = 11.84EE71 pKa = 4.01DD72 pKa = 3.39GKK74 pKa = 9.84EE75 pKa = 3.83TTEE78 pKa = 4.28FEE80 pKa = 4.25TEE82 pKa = 4.07AVVDD86 pKa = 5.15SDD88 pKa = 4.91DD89 pKa = 4.14CGCAYY94 pKa = 9.65EE95 pKa = 4.21YY96 pKa = 10.76HH97 pKa = 6.68SEE99 pKa = 4.31TEE101 pKa = 4.93SEE103 pKa = 4.24ACLCPGYY110 pKa = 10.38AIEE113 pKa = 6.16GICDD117 pKa = 3.66CDD119 pKa = 3.32WYY121 pKa = 10.58EE122 pKa = 4.96DD123 pKa = 3.82RR124 pKa = 11.84PEE126 pKa = 4.2TSDD129 pKa = 3.32SSEE132 pKa = 5.88LFTQWEE138 pKa = 4.12RR139 pKa = 11.84LEE141 pKa = 4.54LFSDD145 pKa = 4.09
MM1 pKa = 7.2SRR3 pKa = 11.84VATIEE8 pKa = 3.82IYY10 pKa = 10.63NEE12 pKa = 3.42NGIIVARR19 pKa = 11.84KK20 pKa = 7.5KK21 pKa = 9.37TSGPHH26 pKa = 6.22ALLEE30 pKa = 4.23LFNGKK35 pKa = 8.18QKK37 pKa = 10.76YY38 pKa = 9.55DD39 pKa = 3.44QVSEE43 pKa = 4.45LFVIWVCEE51 pKa = 4.03EE52 pKa = 4.0CGKK55 pKa = 8.43TVYY58 pKa = 8.96STCEE62 pKa = 3.36FKK64 pKa = 11.22GIVFVRR70 pKa = 11.84EE71 pKa = 4.01DD72 pKa = 3.39GKK74 pKa = 9.84EE75 pKa = 3.83TTEE78 pKa = 4.28FEE80 pKa = 4.25TEE82 pKa = 4.07AVVDD86 pKa = 5.15SDD88 pKa = 4.91DD89 pKa = 4.14CGCAYY94 pKa = 9.65EE95 pKa = 4.21YY96 pKa = 10.76HH97 pKa = 6.68SEE99 pKa = 4.31TEE101 pKa = 4.93SEE103 pKa = 4.24ACLCPGYY110 pKa = 10.38AIEE113 pKa = 6.16GICDD117 pKa = 3.66CDD119 pKa = 3.32WYY121 pKa = 10.58EE122 pKa = 4.96DD123 pKa = 3.82RR124 pKa = 11.84PEE126 pKa = 4.2TSDD129 pKa = 3.32SSEE132 pKa = 5.88LFTQWEE138 pKa = 4.12RR139 pKa = 11.84LEE141 pKa = 4.54LFSDD145 pKa = 4.09
Molecular weight: 16.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4YZ53|S4YZ53_9VIRU N-terminal protein OS=Turnip rosette virus OX=218923 PE=4 SV=1
MM1 pKa = 7.57EE2 pKa = 5.84KK3 pKa = 10.76GNKK6 pKa = 9.45KK7 pKa = 8.23LTKK10 pKa = 8.21NQRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 7.18TRR17 pKa = 11.84KK18 pKa = 9.4KK19 pKa = 8.98NSQSGRR25 pKa = 11.84TNSVVTPLWAPVSTGTVMQGGALSISSLNNKK56 pKa = 9.39GDD58 pKa = 3.05IRR60 pKa = 11.84VCGRR64 pKa = 11.84EE65 pKa = 3.91VVTEE69 pKa = 3.9ISVSNTPTPSVLQVIPEE86 pKa = 4.52TFPSRR91 pKa = 11.84LKK93 pKa = 11.12GLATSWSKK101 pKa = 10.28FKK103 pKa = 9.86WQAIKK108 pKa = 10.06FVYY111 pKa = 9.75MPICPTTEE119 pKa = 3.63RR120 pKa = 11.84GSVHH124 pKa = 7.23FGFLYY129 pKa = 9.55DD130 pKa = 4.11TVDD133 pKa = 3.64NLPGTVGEE141 pKa = 4.18ISTLQGYY148 pKa = 6.09TTGSVWAGTSGNEE161 pKa = 3.88LLEE164 pKa = 5.16DD165 pKa = 3.52GVYY168 pKa = 11.03AKK170 pKa = 9.69TPKK173 pKa = 10.15DD174 pKa = 3.38AVVARR179 pKa = 11.84MDD181 pKa = 4.02ARR183 pKa = 11.84RR184 pKa = 11.84ADD186 pKa = 3.28KK187 pKa = 10.27KK188 pKa = 10.65YY189 pKa = 10.12YY190 pKa = 9.64PIVSTTQLTKK200 pKa = 10.76SLNVDD205 pKa = 3.11ASLGNTYY212 pKa = 10.23VPARR216 pKa = 11.84LAILTADD223 pKa = 3.7GTTAADD229 pKa = 3.57KK230 pKa = 9.88PQIVGRR236 pKa = 11.84LYY238 pKa = 11.1AVFCVDD244 pKa = 6.16LIDD247 pKa = 5.18TIASSLNVV255 pKa = 3.33
MM1 pKa = 7.57EE2 pKa = 5.84KK3 pKa = 10.76GNKK6 pKa = 9.45KK7 pKa = 8.23LTKK10 pKa = 8.21NQRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 7.18TRR17 pKa = 11.84KK18 pKa = 9.4KK19 pKa = 8.98NSQSGRR25 pKa = 11.84TNSVVTPLWAPVSTGTVMQGGALSISSLNNKK56 pKa = 9.39GDD58 pKa = 3.05IRR60 pKa = 11.84VCGRR64 pKa = 11.84EE65 pKa = 3.91VVTEE69 pKa = 3.9ISVSNTPTPSVLQVIPEE86 pKa = 4.52TFPSRR91 pKa = 11.84LKK93 pKa = 11.12GLATSWSKK101 pKa = 10.28FKK103 pKa = 9.86WQAIKK108 pKa = 10.06FVYY111 pKa = 9.75MPICPTTEE119 pKa = 3.63RR120 pKa = 11.84GSVHH124 pKa = 7.23FGFLYY129 pKa = 9.55DD130 pKa = 4.11TVDD133 pKa = 3.64NLPGTVGEE141 pKa = 4.18ISTLQGYY148 pKa = 6.09TTGSVWAGTSGNEE161 pKa = 3.88LLEE164 pKa = 5.16DD165 pKa = 3.52GVYY168 pKa = 11.03AKK170 pKa = 9.69TPKK173 pKa = 10.15DD174 pKa = 3.38AVVARR179 pKa = 11.84MDD181 pKa = 4.02ARR183 pKa = 11.84RR184 pKa = 11.84ADD186 pKa = 3.28KK187 pKa = 10.27KK188 pKa = 10.65YY189 pKa = 10.12YY190 pKa = 9.64PIVSTTQLTKK200 pKa = 10.76SLNVDD205 pKa = 3.11ASLGNTYY212 pKa = 10.23VPARR216 pKa = 11.84LAILTADD223 pKa = 3.7GTTAADD229 pKa = 3.57KK230 pKa = 9.88PQIVGRR236 pKa = 11.84LYY238 pKa = 11.1AVFCVDD244 pKa = 6.16LIDD247 pKa = 5.18TIASSLNVV255 pKa = 3.33
Molecular weight: 27.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1867 |
145 |
929 |
466.8 |
51.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.999 ± 0.23 | 1.768 ± 0.715 |
4.66 ± 0.383 | 7.445 ± 1.404 |
3.16 ± 0.491 | 7.177 ± 0.169 |
1.821 ± 0.304 | 4.928 ± 0.103 |
5.785 ± 0.265 | 8.784 ± 0.617 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.5 ± 0.134 | 3.642 ± 0.267 |
4.66 ± 0.411 | 2.946 ± 0.211 |
5.731 ± 0.431 | 9.373 ± 0.758 |
7.177 ± 1.04 | 8.784 ± 0.306 |
1.875 ± 0.196 | 2.785 ± 0.428 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
