
Panagrellus redivivus (Microworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Tylenchina; Panagrolaimomorpha; Panagrolaimoidea; Panagrolaimidae; Panagrellus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
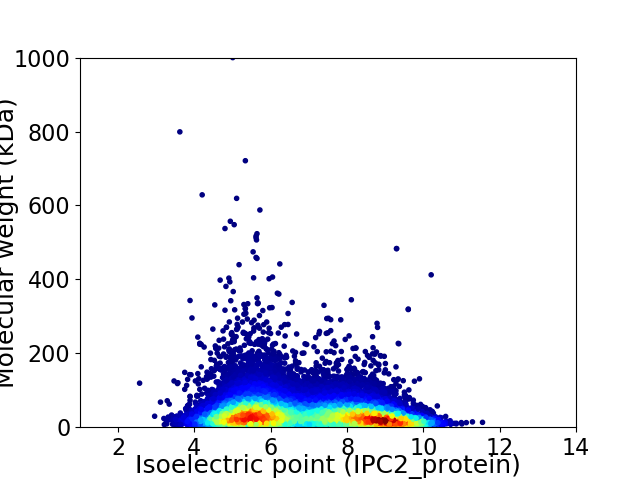
Virtual 2D-PAGE plot for 26326 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
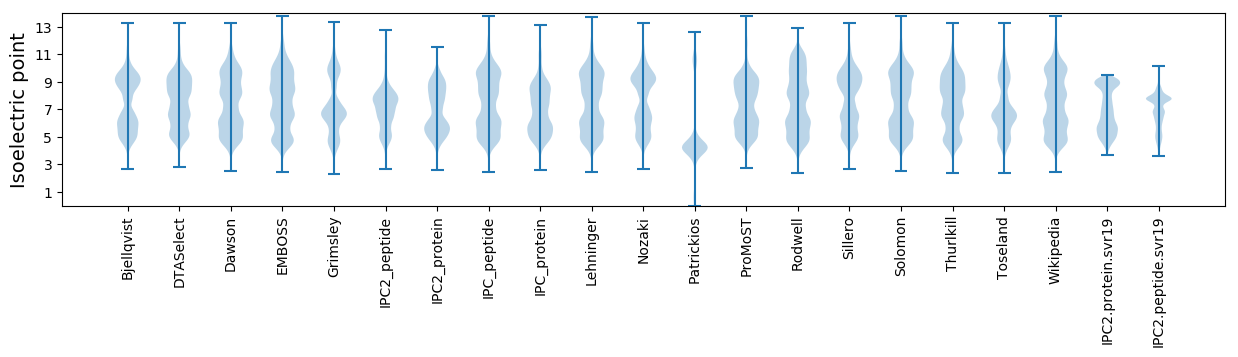
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7E4VBK4|A0A7E4VBK4_PANRE Uncharacterized protein OS=Panagrellus redivivus OX=6233 PE=4 SV=1
MM1 pKa = 7.62SNRR4 pKa = 11.84SIKK7 pKa = 9.23TLNEE11 pKa = 3.65AEE13 pKa = 4.62EE14 pKa = 4.17VHH16 pKa = 6.23SLYY19 pKa = 10.91VFQDD23 pKa = 3.05TDD25 pKa = 3.96DD26 pKa = 4.76DD27 pKa = 4.29VASVYY32 pKa = 11.13AFDD35 pKa = 5.09APSKK39 pKa = 10.19ICFKK43 pKa = 10.7GSSSAYY49 pKa = 10.03AFWSTLFFSVWAYY62 pKa = 9.19LQYY65 pKa = 11.06VIAYY69 pKa = 8.87LYY71 pKa = 10.8QIDD74 pKa = 4.14NNSIIAEE81 pKa = 4.37TVNSNLNSEE90 pKa = 4.1VDD92 pKa = 3.35TLKK95 pKa = 11.1ANKK98 pKa = 9.99ADD100 pKa = 4.19DD101 pKa = 4.72DD102 pKa = 4.59SVSVYY107 pKa = 11.07AFDD110 pKa = 5.29NSDD113 pKa = 3.8SPSTDD118 pKa = 2.99ADD120 pKa = 3.8KK121 pKa = 10.5DD122 pKa = 3.72TDD124 pKa = 3.99DD125 pKa = 4.15VEE127 pKa = 4.54SVYY130 pKa = 11.27AFDD133 pKa = 5.37GADD136 pKa = 3.24NDD138 pKa = 4.82EE139 pKa = 4.6IQSSDD144 pKa = 3.51DD145 pKa = 3.62TEE147 pKa = 5.19GSVSSEE153 pKa = 3.84SSNSFGFTIPNLGEE167 pKa = 4.13GEE169 pKa = 4.25QYY171 pKa = 10.97GYY173 pKa = 10.6HH174 pKa = 6.97ADD176 pKa = 3.66SDD178 pKa = 4.09DD179 pKa = 3.84EE180 pKa = 5.95RR181 pKa = 11.84FDD183 pKa = 3.85FYY185 pKa = 11.96NNVTCVRR192 pKa = 11.84LVTGEE197 pKa = 4.24SLTLAFDD204 pKa = 3.89GQNLKK209 pKa = 10.64VVIHH213 pKa = 7.06DD214 pKa = 4.02GKK216 pKa = 10.96AYY218 pKa = 9.7TSFDD222 pKa = 3.49AMPSDD227 pKa = 4.41VADD230 pKa = 4.11LLIAARR236 pKa = 11.84ARR238 pKa = 11.84IIDD241 pKa = 3.76LVFF244 pKa = 4.55
MM1 pKa = 7.62SNRR4 pKa = 11.84SIKK7 pKa = 9.23TLNEE11 pKa = 3.65AEE13 pKa = 4.62EE14 pKa = 4.17VHH16 pKa = 6.23SLYY19 pKa = 10.91VFQDD23 pKa = 3.05TDD25 pKa = 3.96DD26 pKa = 4.76DD27 pKa = 4.29VASVYY32 pKa = 11.13AFDD35 pKa = 5.09APSKK39 pKa = 10.19ICFKK43 pKa = 10.7GSSSAYY49 pKa = 10.03AFWSTLFFSVWAYY62 pKa = 9.19LQYY65 pKa = 11.06VIAYY69 pKa = 8.87LYY71 pKa = 10.8QIDD74 pKa = 4.14NNSIIAEE81 pKa = 4.37TVNSNLNSEE90 pKa = 4.1VDD92 pKa = 3.35TLKK95 pKa = 11.1ANKK98 pKa = 9.99ADD100 pKa = 4.19DD101 pKa = 4.72DD102 pKa = 4.59SVSVYY107 pKa = 11.07AFDD110 pKa = 5.29NSDD113 pKa = 3.8SPSTDD118 pKa = 2.99ADD120 pKa = 3.8KK121 pKa = 10.5DD122 pKa = 3.72TDD124 pKa = 3.99DD125 pKa = 4.15VEE127 pKa = 4.54SVYY130 pKa = 11.27AFDD133 pKa = 5.37GADD136 pKa = 3.24NDD138 pKa = 4.82EE139 pKa = 4.6IQSSDD144 pKa = 3.51DD145 pKa = 3.62TEE147 pKa = 5.19GSVSSEE153 pKa = 3.84SSNSFGFTIPNLGEE167 pKa = 4.13GEE169 pKa = 4.25QYY171 pKa = 10.97GYY173 pKa = 10.6HH174 pKa = 6.97ADD176 pKa = 3.66SDD178 pKa = 4.09DD179 pKa = 3.84EE180 pKa = 5.95RR181 pKa = 11.84FDD183 pKa = 3.85FYY185 pKa = 11.96NNVTCVRR192 pKa = 11.84LVTGEE197 pKa = 4.24SLTLAFDD204 pKa = 3.89GQNLKK209 pKa = 10.64VVIHH213 pKa = 7.06DD214 pKa = 4.02GKK216 pKa = 10.96AYY218 pKa = 9.7TSFDD222 pKa = 3.49AMPSDD227 pKa = 4.41VADD230 pKa = 4.11LLIAARR236 pKa = 11.84ARR238 pKa = 11.84IIDD241 pKa = 3.76LVFF244 pKa = 4.55
Molecular weight: 26.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7E4VW86|A0A7E4VW86_PANRE Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit KCP2 OS=Panagrellus redivivus OX=6233 PE=3 SV=1
AA1 pKa = 7.85RR2 pKa = 11.84PLASNQPPARR12 pKa = 11.84QPKK15 pKa = 9.37ARR17 pKa = 11.84HH18 pKa = 5.32QGTVRR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84APLKK30 pKa = 10.56ARR32 pKa = 11.84PLASNQPPARR42 pKa = 11.84QPKK45 pKa = 8.68ARR47 pKa = 11.84HH48 pKa = 5.99RR49 pKa = 11.84GTARR53 pKa = 11.84RR54 pKa = 11.84PKK56 pKa = 10.42APLKK60 pKa = 10.51ARR62 pKa = 11.84PLASNQPPARR72 pKa = 11.84QPKK75 pKa = 9.44ARR77 pKa = 11.84PLATNPPPARR87 pKa = 11.84QPKK90 pKa = 9.41ARR92 pKa = 11.84PLASNQPPARR102 pKa = 11.84QPKK105 pKa = 9.32ARR107 pKa = 11.84HH108 pKa = 5.95RR109 pKa = 11.84DD110 pKa = 3.16SARR113 pKa = 11.84RR114 pKa = 11.84PRR116 pKa = 11.84APLKK120 pKa = 10.56ARR122 pKa = 11.84PLASNQPPAA131 pKa = 4.28
AA1 pKa = 7.85RR2 pKa = 11.84PLASNQPPARR12 pKa = 11.84QPKK15 pKa = 9.37ARR17 pKa = 11.84HH18 pKa = 5.32QGTVRR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84APLKK30 pKa = 10.56ARR32 pKa = 11.84PLASNQPPARR42 pKa = 11.84QPKK45 pKa = 8.68ARR47 pKa = 11.84HH48 pKa = 5.99RR49 pKa = 11.84GTARR53 pKa = 11.84RR54 pKa = 11.84PKK56 pKa = 10.42APLKK60 pKa = 10.51ARR62 pKa = 11.84PLASNQPPARR72 pKa = 11.84QPKK75 pKa = 9.44ARR77 pKa = 11.84PLATNPPPARR87 pKa = 11.84QPKK90 pKa = 9.41ARR92 pKa = 11.84PLASNQPPARR102 pKa = 11.84QPKK105 pKa = 9.32ARR107 pKa = 11.84HH108 pKa = 5.95RR109 pKa = 11.84DD110 pKa = 3.16SARR113 pKa = 11.84RR114 pKa = 11.84PRR116 pKa = 11.84APLKK120 pKa = 10.56ARR122 pKa = 11.84PLASNQPPAA131 pKa = 4.28
Molecular weight: 14.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10071718 |
11 |
9598 |
382.6 |
42.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.581 ± 0.024 | 1.958 ± 0.015 |
5.584 ± 0.015 | 5.902 ± 0.024 |
4.453 ± 0.015 | 5.408 ± 0.02 |
2.399 ± 0.008 | 5.494 ± 0.015 |
5.712 ± 0.022 | 8.472 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.317 ± 0.008 | 4.668 ± 0.012 |
5.827 ± 0.02 | 3.772 ± 0.016 |
5.468 ± 0.017 | 7.734 ± 0.022 |
6.257 ± 0.021 | 6.646 ± 0.012 |
1.02 ± 0.005 | 3.138 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
