
Agrococcus jejuensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agrococcus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
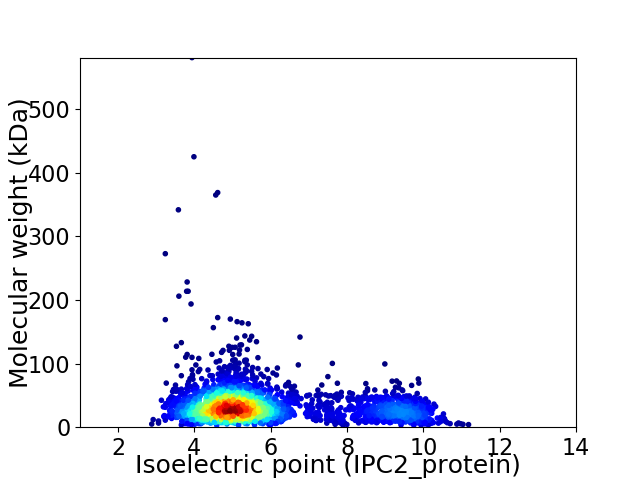
Virtual 2D-PAGE plot for 3218 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
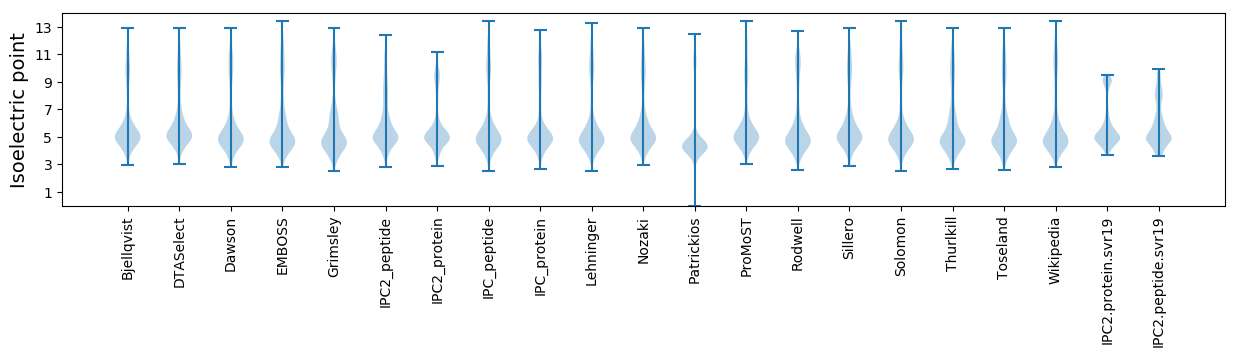
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8A2I9|A0A1G8A2I9_9MICO FtsX-like permease family protein OS=Agrococcus jejuensis OX=399736 GN=SAMN04489720_0223 PE=4 SV=1
MM1 pKa = 8.19RR2 pKa = 11.84IRR4 pKa = 11.84PAVLVLAASVSLVGCTASDD23 pKa = 3.45PAPRR27 pKa = 11.84VTADD31 pKa = 3.51DD32 pKa = 3.89VADD35 pKa = 4.33RR36 pKa = 11.84WAQASQGAIDD46 pKa = 3.98YY47 pKa = 10.09QLGGAYY53 pKa = 10.06DD54 pKa = 4.06PAPGTTIVARR64 pKa = 11.84DD65 pKa = 3.58ASDD68 pKa = 3.92PPAEE72 pKa = 4.26GAFSICYY79 pKa = 10.32LNGFQTQPADD89 pKa = 3.84GEE91 pKa = 4.33WWLEE95 pKa = 3.82EE96 pKa = 4.27HH97 pKa = 7.31PDD99 pKa = 4.04LVLRR103 pKa = 11.84DD104 pKa = 3.81AAGEE108 pKa = 3.93PAIDD112 pKa = 4.38PGWPDD117 pKa = 3.67EE118 pKa = 4.2MALDD122 pKa = 4.07TSTPEE127 pKa = 3.62QRR129 pKa = 11.84EE130 pKa = 3.85QIAAIVTPWIAGCASDD146 pKa = 5.88GYY148 pKa = 11.27DD149 pKa = 3.73AVEE152 pKa = 4.44FDD154 pKa = 5.87NLDD157 pKa = 2.81SWTRR161 pKa = 11.84FDD163 pKa = 3.82GLTMEE168 pKa = 5.26GNMALAALLVDD179 pKa = 4.28AAHH182 pKa = 7.9DD183 pKa = 3.84EE184 pKa = 4.56GLWAAQKK191 pKa = 10.4NALDD195 pKa = 4.25AGEE198 pKa = 4.83AGPGAGFDD206 pKa = 3.49LVVTEE211 pKa = 4.41EE212 pKa = 4.01CGQYY216 pKa = 9.96EE217 pKa = 4.24EE218 pKa = 4.59CAAYY222 pKa = 10.35AEE224 pKa = 4.9LYY226 pKa = 10.33GDD228 pKa = 3.31RR229 pKa = 11.84HH230 pKa = 6.71VDD232 pKa = 3.01IEE234 pKa = 4.57YY235 pKa = 10.9VEE237 pKa = 4.46STSEE241 pKa = 3.85DD242 pKa = 3.55DD243 pKa = 3.54FAAMCEE249 pKa = 4.25AGDD252 pKa = 3.98VPPLTVLRR260 pKa = 11.84DD261 pKa = 3.46RR262 pKa = 11.84PLSVEE267 pKa = 3.86GDD269 pKa = 3.75DD270 pKa = 3.54GHH272 pKa = 7.94VRR274 pKa = 11.84VPCSS278 pKa = 2.98
MM1 pKa = 8.19RR2 pKa = 11.84IRR4 pKa = 11.84PAVLVLAASVSLVGCTASDD23 pKa = 3.45PAPRR27 pKa = 11.84VTADD31 pKa = 3.51DD32 pKa = 3.89VADD35 pKa = 4.33RR36 pKa = 11.84WAQASQGAIDD46 pKa = 3.98YY47 pKa = 10.09QLGGAYY53 pKa = 10.06DD54 pKa = 4.06PAPGTTIVARR64 pKa = 11.84DD65 pKa = 3.58ASDD68 pKa = 3.92PPAEE72 pKa = 4.26GAFSICYY79 pKa = 10.32LNGFQTQPADD89 pKa = 3.84GEE91 pKa = 4.33WWLEE95 pKa = 3.82EE96 pKa = 4.27HH97 pKa = 7.31PDD99 pKa = 4.04LVLRR103 pKa = 11.84DD104 pKa = 3.81AAGEE108 pKa = 3.93PAIDD112 pKa = 4.38PGWPDD117 pKa = 3.67EE118 pKa = 4.2MALDD122 pKa = 4.07TSTPEE127 pKa = 3.62QRR129 pKa = 11.84EE130 pKa = 3.85QIAAIVTPWIAGCASDD146 pKa = 5.88GYY148 pKa = 11.27DD149 pKa = 3.73AVEE152 pKa = 4.44FDD154 pKa = 5.87NLDD157 pKa = 2.81SWTRR161 pKa = 11.84FDD163 pKa = 3.82GLTMEE168 pKa = 5.26GNMALAALLVDD179 pKa = 4.28AAHH182 pKa = 7.9DD183 pKa = 3.84EE184 pKa = 4.56GLWAAQKK191 pKa = 10.4NALDD195 pKa = 4.25AGEE198 pKa = 4.83AGPGAGFDD206 pKa = 3.49LVVTEE211 pKa = 4.41EE212 pKa = 4.01CGQYY216 pKa = 9.96EE217 pKa = 4.24EE218 pKa = 4.59CAAYY222 pKa = 10.35AEE224 pKa = 4.9LYY226 pKa = 10.33GDD228 pKa = 3.31RR229 pKa = 11.84HH230 pKa = 6.71VDD232 pKa = 3.01IEE234 pKa = 4.57YY235 pKa = 10.9VEE237 pKa = 4.46STSEE241 pKa = 3.85DD242 pKa = 3.55DD243 pKa = 3.54FAAMCEE249 pKa = 4.25AGDD252 pKa = 3.98VPPLTVLRR260 pKa = 11.84DD261 pKa = 3.46RR262 pKa = 11.84PLSVEE267 pKa = 3.86GDD269 pKa = 3.75DD270 pKa = 3.54GHH272 pKa = 7.94VRR274 pKa = 11.84VPCSS278 pKa = 2.98
Molecular weight: 29.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8EVG8|A0A1G8EVG8_9MICO T5orf172 domain-containing protein OS=Agrococcus jejuensis OX=399736 GN=SAMN04489720_2205 PE=4 SV=1
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NRR31 pKa = 11.84KK32 pKa = 8.65
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NRR31 pKa = 11.84KK32 pKa = 8.65
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1039962 |
29 |
5825 |
323.2 |
34.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.682 ± 0.082 | 0.497 ± 0.01 |
6.632 ± 0.042 | 5.54 ± 0.051 |
2.714 ± 0.026 | 9.104 ± 0.054 |
2.067 ± 0.029 | 4.202 ± 0.03 |
1.366 ± 0.031 | 10.0 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.85 ± 0.023 | 1.432 ± 0.026 |
5.485 ± 0.049 | 2.967 ± 0.034 |
7.339 ± 0.071 | 5.097 ± 0.031 |
6.119 ± 0.086 | 9.753 ± 0.049 |
1.486 ± 0.021 | 1.667 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
