
Paracoccus sp. JM45
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus; unclassified Paracoccus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
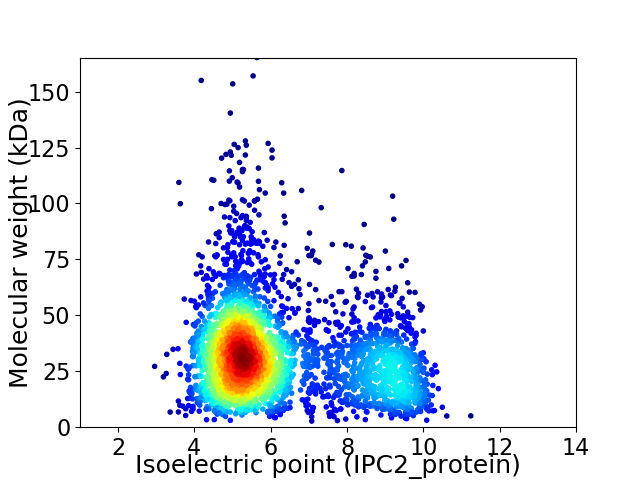
Virtual 2D-PAGE plot for 3379 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
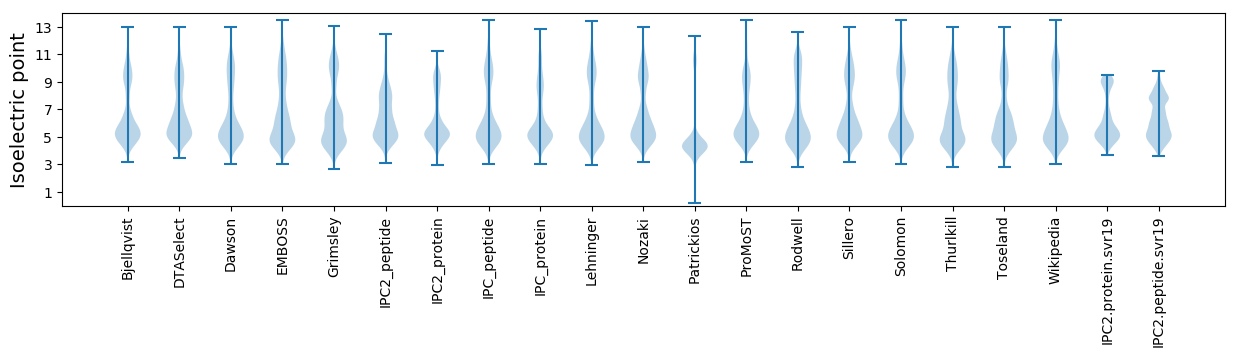
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A3DU69|A0A3A3DU69_9RHOB IS6 family transposase (Fragment) OS=Paracoccus sp. JM45 OX=2283626 GN=DWB67_16910 PE=4 SV=1
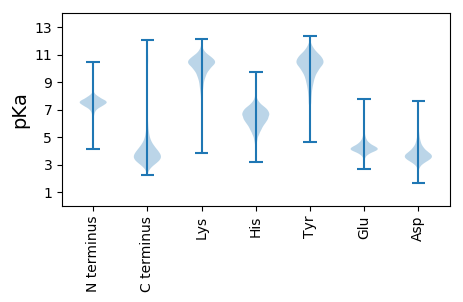
MM1 pKa = 7.58VSTAQLNIVNLQVEE15 pKa = 4.6AAGQIAIDD23 pKa = 4.07KK24 pKa = 11.04ANGSYY29 pKa = 10.4AANLAANDD37 pKa = 5.0KK38 pKa = 10.38YY39 pKa = 11.13YY40 pKa = 11.03LWQSDD45 pKa = 4.09FQTLPTSMFGEE56 pKa = 4.32PEE58 pKa = 4.09LYY60 pKa = 10.47AEE62 pKa = 4.8RR63 pKa = 11.84LHH65 pKa = 7.27DD66 pKa = 4.76ALPGVTTLRR75 pKa = 11.84IPFNLNSFNADD86 pKa = 3.07GTLHH90 pKa = 7.08PDD92 pKa = 3.66FEE94 pKa = 5.09RR95 pKa = 11.84FLVAAANEE103 pKa = 4.31GFTFIMVQMEE113 pKa = 4.57GAAQTLTASGPNALDD128 pKa = 3.22QMRR131 pKa = 11.84DD132 pKa = 3.45GLSVEE137 pKa = 4.45VYY139 pKa = 11.15DD140 pKa = 5.98RR141 pKa = 11.84MEE143 pKa = 4.18QGWTMLLDD151 pKa = 3.47WMDD154 pKa = 3.36THH156 pKa = 7.1PSVKK160 pKa = 10.21DD161 pKa = 3.01SVYY164 pKa = 9.78AHH166 pKa = 5.99EE167 pKa = 4.83VVNEE171 pKa = 3.61PAAYY175 pKa = 7.98NTATFYY181 pKa = 9.73GTSRR185 pKa = 11.84PAALQDD191 pKa = 3.91FVSLYY196 pKa = 8.23ATHH199 pKa = 6.66MVQLGQMIQDD209 pKa = 3.58RR210 pKa = 11.84ADD212 pKa = 3.27GTKK215 pKa = 10.34IMVGGWNYY223 pKa = 10.66SAQFQQLADD232 pKa = 3.18ISMGGGSALDD242 pKa = 3.97YY243 pKa = 10.78IRR245 pKa = 11.84EE246 pKa = 4.16GLGDD250 pKa = 3.79SLVWSAHH257 pKa = 6.82LYY259 pKa = 9.78PGWLGTGGMSDD270 pKa = 3.61PNAIRR275 pKa = 11.84TVLDD279 pKa = 4.37EE280 pKa = 4.68IYY282 pKa = 10.97SSIVDD287 pKa = 3.66DD288 pKa = 4.69SLILSEE294 pKa = 4.85TNAQGNEE301 pKa = 3.77AYY303 pKa = 10.72NLFSDD308 pKa = 3.9RR309 pKa = 11.84PEE311 pKa = 4.02VQGFTQVYY319 pKa = 9.97DD320 pKa = 3.31WFADD324 pKa = 3.68RR325 pKa = 11.84GIGISWFTGSQYY337 pKa = 11.15GGSVLSRR344 pKa = 11.84MDD346 pKa = 3.76PDD348 pKa = 3.6GTLSFVQQGSFGAAMDD364 pKa = 4.25AFTLGGEE371 pKa = 4.57DD372 pKa = 3.99PDD374 pKa = 4.08HH375 pKa = 7.05AGGEE379 pKa = 4.38VVEE382 pKa = 4.34IQLIRR387 pKa = 11.84GRR389 pKa = 11.84MRR391 pKa = 11.84NQTTDD396 pKa = 3.38PDD398 pKa = 3.73YY399 pKa = 10.88QAYY402 pKa = 10.83NEE404 pKa = 3.94MDD406 pKa = 2.9IAQFLGSGLGHH417 pKa = 7.11GGNDD421 pKa = 3.57TLTGSAVANNFLYY434 pKa = 10.76GGTSNDD440 pKa = 3.4LVTGNILDD448 pKa = 4.24DD449 pKa = 3.95FLYY452 pKa = 10.75GQDD455 pKa = 4.88DD456 pKa = 4.22NDD458 pKa = 4.6TIDD461 pKa = 4.24GGLSGHH467 pKa = 6.34DD468 pKa = 3.46HH469 pKa = 6.75LFGGGGEE476 pKa = 4.2DD477 pKa = 5.38LIIGGAGISQMYY489 pKa = 9.74GGTGGDD495 pKa = 3.27TFVAHH500 pKa = 6.8PRR502 pKa = 11.84GQTIIVDD509 pKa = 3.65YY510 pKa = 10.92DD511 pKa = 3.64STEE514 pKa = 3.9GDD516 pKa = 3.22QLIINEE522 pKa = 4.21ADD524 pKa = 3.53FTVEE528 pKa = 3.98DD529 pKa = 3.87VRR531 pKa = 11.84RR532 pKa = 11.84LAQSLDD538 pKa = 2.82WDD540 pKa = 4.48LDD542 pKa = 3.53DD543 pKa = 6.52SRR545 pKa = 11.84DD546 pKa = 3.98LRR548 pKa = 11.84IALPGGGEE556 pKa = 4.09IIMLGMGDD564 pKa = 3.57RR565 pKa = 11.84LEE567 pKa = 5.04DD568 pKa = 3.66VIASLFPAQGNVEE581 pKa = 4.18PAAPAPFHH589 pKa = 6.8LLLQGRR595 pKa = 11.84PGVDD599 pKa = 2.74ILAGGAGNDD608 pKa = 4.01TIYY611 pKa = 11.0GDD613 pKa = 3.62PEE615 pKa = 4.24GAVGADD621 pKa = 3.37QLYY624 pKa = 10.92GGANRR629 pKa = 11.84DD630 pKa = 3.79LIYY633 pKa = 11.0GGGEE637 pKa = 3.77ADD639 pKa = 4.44YY640 pKa = 11.16IDD642 pKa = 5.13GGTGNDD648 pKa = 3.31TLYY651 pKa = 11.11GGEE654 pKa = 4.52GGDD657 pKa = 4.06AIHH660 pKa = 7.1GGLHH664 pKa = 6.21NDD666 pKa = 3.12VLEE669 pKa = 4.78GGGGDD674 pKa = 3.82DD675 pKa = 4.13TVHH678 pKa = 6.61GSNGQDD684 pKa = 3.34VLSGGLGSDD693 pKa = 3.59TLSGDD698 pKa = 3.51GGRR701 pKa = 11.84DD702 pKa = 3.12TLYY705 pKa = 11.23GGDD708 pKa = 4.4SSDD711 pKa = 4.04LLLGGGGNDD720 pKa = 4.02MIFGGNGNDD729 pKa = 3.31TLYY732 pKa = 11.26GGNRR736 pKa = 11.84NDD738 pKa = 4.83LLYY741 pKa = 11.15GDD743 pKa = 5.01ASNDD747 pKa = 3.29VLYY750 pKa = 11.03GEE752 pKa = 4.74KK753 pKa = 10.2WNDD756 pKa = 3.26TLYY759 pKa = 11.33GGAGQDD765 pKa = 3.53DD766 pKa = 5.51LYY768 pKa = 11.52GGTEE772 pKa = 4.1DD773 pKa = 5.23DD774 pKa = 5.27DD775 pKa = 5.95LYY777 pKa = 11.71GEE779 pKa = 4.52NSNDD783 pKa = 3.66TLDD786 pKa = 4.12GGSGNDD792 pKa = 3.45MLDD795 pKa = 3.31GGGQNDD801 pKa = 4.64LLYY804 pKa = 11.01GGTGNDD810 pKa = 3.98DD811 pKa = 3.38LHH813 pKa = 8.58GGFGLDD819 pKa = 3.08TLYY822 pKa = 11.41GEE824 pKa = 5.33NGDD827 pKa = 3.96EE828 pKa = 4.17LLNGGGMADD837 pKa = 4.06MIYY840 pKa = 10.56GGSGNDD846 pKa = 3.51TLLGEE851 pKa = 5.13AGNDD855 pKa = 3.97WIHH858 pKa = 6.6GGPGQDD864 pKa = 3.1VATGGGGADD873 pKa = 3.1TFVFDD878 pKa = 5.61LGDD881 pKa = 3.68GSLRR885 pKa = 11.84ITDD888 pKa = 4.14FSVNQGDD895 pKa = 4.41ILRR898 pKa = 11.84LDD900 pKa = 3.53AALLDD905 pKa = 4.37GAPSVAALQEE915 pKa = 4.31RR916 pKa = 11.84ATLTDD921 pKa = 3.57AGTSIIFTDD930 pKa = 3.99GNTVLIANFQGTFSDD945 pKa = 3.75SFVDD949 pKa = 3.79ILL951 pKa = 4.53
MM1 pKa = 7.58VSTAQLNIVNLQVEE15 pKa = 4.6AAGQIAIDD23 pKa = 4.07KK24 pKa = 11.04ANGSYY29 pKa = 10.4AANLAANDD37 pKa = 5.0KK38 pKa = 10.38YY39 pKa = 11.13YY40 pKa = 11.03LWQSDD45 pKa = 4.09FQTLPTSMFGEE56 pKa = 4.32PEE58 pKa = 4.09LYY60 pKa = 10.47AEE62 pKa = 4.8RR63 pKa = 11.84LHH65 pKa = 7.27DD66 pKa = 4.76ALPGVTTLRR75 pKa = 11.84IPFNLNSFNADD86 pKa = 3.07GTLHH90 pKa = 7.08PDD92 pKa = 3.66FEE94 pKa = 5.09RR95 pKa = 11.84FLVAAANEE103 pKa = 4.31GFTFIMVQMEE113 pKa = 4.57GAAQTLTASGPNALDD128 pKa = 3.22QMRR131 pKa = 11.84DD132 pKa = 3.45GLSVEE137 pKa = 4.45VYY139 pKa = 11.15DD140 pKa = 5.98RR141 pKa = 11.84MEE143 pKa = 4.18QGWTMLLDD151 pKa = 3.47WMDD154 pKa = 3.36THH156 pKa = 7.1PSVKK160 pKa = 10.21DD161 pKa = 3.01SVYY164 pKa = 9.78AHH166 pKa = 5.99EE167 pKa = 4.83VVNEE171 pKa = 3.61PAAYY175 pKa = 7.98NTATFYY181 pKa = 9.73GTSRR185 pKa = 11.84PAALQDD191 pKa = 3.91FVSLYY196 pKa = 8.23ATHH199 pKa = 6.66MVQLGQMIQDD209 pKa = 3.58RR210 pKa = 11.84ADD212 pKa = 3.27GTKK215 pKa = 10.34IMVGGWNYY223 pKa = 10.66SAQFQQLADD232 pKa = 3.18ISMGGGSALDD242 pKa = 3.97YY243 pKa = 10.78IRR245 pKa = 11.84EE246 pKa = 4.16GLGDD250 pKa = 3.79SLVWSAHH257 pKa = 6.82LYY259 pKa = 9.78PGWLGTGGMSDD270 pKa = 3.61PNAIRR275 pKa = 11.84TVLDD279 pKa = 4.37EE280 pKa = 4.68IYY282 pKa = 10.97SSIVDD287 pKa = 3.66DD288 pKa = 4.69SLILSEE294 pKa = 4.85TNAQGNEE301 pKa = 3.77AYY303 pKa = 10.72NLFSDD308 pKa = 3.9RR309 pKa = 11.84PEE311 pKa = 4.02VQGFTQVYY319 pKa = 9.97DD320 pKa = 3.31WFADD324 pKa = 3.68RR325 pKa = 11.84GIGISWFTGSQYY337 pKa = 11.15GGSVLSRR344 pKa = 11.84MDD346 pKa = 3.76PDD348 pKa = 3.6GTLSFVQQGSFGAAMDD364 pKa = 4.25AFTLGGEE371 pKa = 4.57DD372 pKa = 3.99PDD374 pKa = 4.08HH375 pKa = 7.05AGGEE379 pKa = 4.38VVEE382 pKa = 4.34IQLIRR387 pKa = 11.84GRR389 pKa = 11.84MRR391 pKa = 11.84NQTTDD396 pKa = 3.38PDD398 pKa = 3.73YY399 pKa = 10.88QAYY402 pKa = 10.83NEE404 pKa = 3.94MDD406 pKa = 2.9IAQFLGSGLGHH417 pKa = 7.11GGNDD421 pKa = 3.57TLTGSAVANNFLYY434 pKa = 10.76GGTSNDD440 pKa = 3.4LVTGNILDD448 pKa = 4.24DD449 pKa = 3.95FLYY452 pKa = 10.75GQDD455 pKa = 4.88DD456 pKa = 4.22NDD458 pKa = 4.6TIDD461 pKa = 4.24GGLSGHH467 pKa = 6.34DD468 pKa = 3.46HH469 pKa = 6.75LFGGGGEE476 pKa = 4.2DD477 pKa = 5.38LIIGGAGISQMYY489 pKa = 9.74GGTGGDD495 pKa = 3.27TFVAHH500 pKa = 6.8PRR502 pKa = 11.84GQTIIVDD509 pKa = 3.65YY510 pKa = 10.92DD511 pKa = 3.64STEE514 pKa = 3.9GDD516 pKa = 3.22QLIINEE522 pKa = 4.21ADD524 pKa = 3.53FTVEE528 pKa = 3.98DD529 pKa = 3.87VRR531 pKa = 11.84RR532 pKa = 11.84LAQSLDD538 pKa = 2.82WDD540 pKa = 4.48LDD542 pKa = 3.53DD543 pKa = 6.52SRR545 pKa = 11.84DD546 pKa = 3.98LRR548 pKa = 11.84IALPGGGEE556 pKa = 4.09IIMLGMGDD564 pKa = 3.57RR565 pKa = 11.84LEE567 pKa = 5.04DD568 pKa = 3.66VIASLFPAQGNVEE581 pKa = 4.18PAAPAPFHH589 pKa = 6.8LLLQGRR595 pKa = 11.84PGVDD599 pKa = 2.74ILAGGAGNDD608 pKa = 4.01TIYY611 pKa = 11.0GDD613 pKa = 3.62PEE615 pKa = 4.24GAVGADD621 pKa = 3.37QLYY624 pKa = 10.92GGANRR629 pKa = 11.84DD630 pKa = 3.79LIYY633 pKa = 11.0GGGEE637 pKa = 3.77ADD639 pKa = 4.44YY640 pKa = 11.16IDD642 pKa = 5.13GGTGNDD648 pKa = 3.31TLYY651 pKa = 11.11GGEE654 pKa = 4.52GGDD657 pKa = 4.06AIHH660 pKa = 7.1GGLHH664 pKa = 6.21NDD666 pKa = 3.12VLEE669 pKa = 4.78GGGGDD674 pKa = 3.82DD675 pKa = 4.13TVHH678 pKa = 6.61GSNGQDD684 pKa = 3.34VLSGGLGSDD693 pKa = 3.59TLSGDD698 pKa = 3.51GGRR701 pKa = 11.84DD702 pKa = 3.12TLYY705 pKa = 11.23GGDD708 pKa = 4.4SSDD711 pKa = 4.04LLLGGGGNDD720 pKa = 4.02MIFGGNGNDD729 pKa = 3.31TLYY732 pKa = 11.26GGNRR736 pKa = 11.84NDD738 pKa = 4.83LLYY741 pKa = 11.15GDD743 pKa = 5.01ASNDD747 pKa = 3.29VLYY750 pKa = 11.03GEE752 pKa = 4.74KK753 pKa = 10.2WNDD756 pKa = 3.26TLYY759 pKa = 11.33GGAGQDD765 pKa = 3.53DD766 pKa = 5.51LYY768 pKa = 11.52GGTEE772 pKa = 4.1DD773 pKa = 5.23DD774 pKa = 5.27DD775 pKa = 5.95LYY777 pKa = 11.71GEE779 pKa = 4.52NSNDD783 pKa = 3.66TLDD786 pKa = 4.12GGSGNDD792 pKa = 3.45MLDD795 pKa = 3.31GGGQNDD801 pKa = 4.64LLYY804 pKa = 11.01GGTGNDD810 pKa = 3.98DD811 pKa = 3.38LHH813 pKa = 8.58GGFGLDD819 pKa = 3.08TLYY822 pKa = 11.41GEE824 pKa = 5.33NGDD827 pKa = 3.96EE828 pKa = 4.17LLNGGGMADD837 pKa = 4.06MIYY840 pKa = 10.56GGSGNDD846 pKa = 3.51TLLGEE851 pKa = 5.13AGNDD855 pKa = 3.97WIHH858 pKa = 6.6GGPGQDD864 pKa = 3.1VATGGGGADD873 pKa = 3.1TFVFDD878 pKa = 5.61LGDD881 pKa = 3.68GSLRR885 pKa = 11.84ITDD888 pKa = 4.14FSVNQGDD895 pKa = 4.41ILRR898 pKa = 11.84LDD900 pKa = 3.53AALLDD905 pKa = 4.37GAPSVAALQEE915 pKa = 4.31RR916 pKa = 11.84ATLTDD921 pKa = 3.57AGTSIIFTDD930 pKa = 3.99GNTVLIANFQGTFSDD945 pKa = 3.75SFVDD949 pKa = 3.79ILL951 pKa = 4.53
Molecular weight: 99.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A3DD12|A0A3A3DD12_9RHOB ABC transporter substrate-binding protein OS=Paracoccus sp. JM45 OX=2283626 GN=DWB67_11230 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.99GGRR28 pKa = 11.84RR29 pKa = 11.84ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.99GGRR28 pKa = 11.84RR29 pKa = 11.84ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1057785 |
26 |
1512 |
313.0 |
34.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.094 ± 0.055 | 0.867 ± 0.011 |
6.442 ± 0.04 | 5.022 ± 0.031 |
3.644 ± 0.026 | 8.552 ± 0.043 |
2.111 ± 0.022 | 5.506 ± 0.034 |
2.967 ± 0.029 | 10.098 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.032 ± 0.022 | 2.69 ± 0.023 |
5.126 ± 0.03 | 3.664 ± 0.022 |
6.709 ± 0.045 | 5.254 ± 0.029 |
5.594 ± 0.026 | 7.076 ± 0.034 |
1.442 ± 0.018 | 2.111 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
