
Gordonia phage PhrostedPhlake
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Fairfaxidumvirus; unclassified Fairfaxidumvirus
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
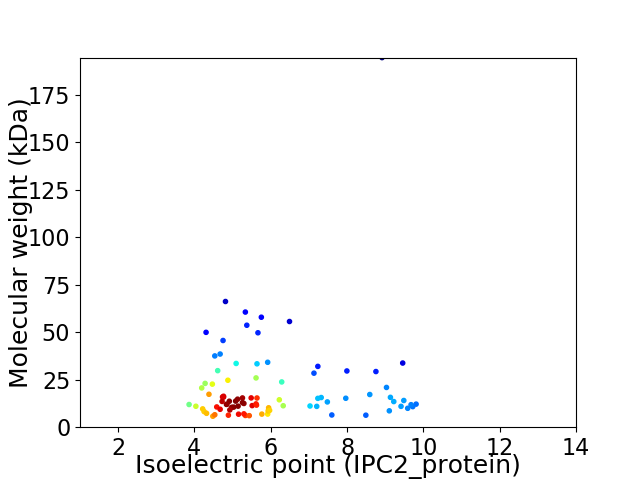
Virtual 2D-PAGE plot for 88 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
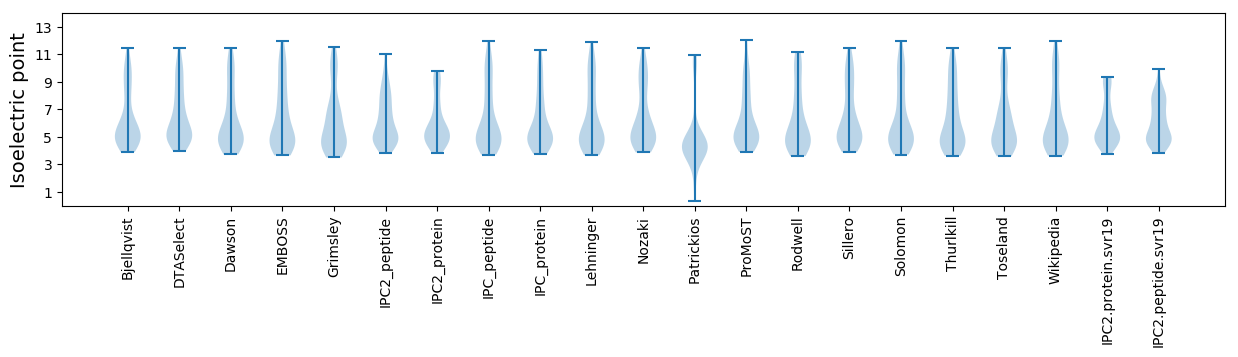
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y6EHI8|A0A4Y6EHI8_9CAUD Helix-turn-helix DNA binding protein OS=Gordonia phage PhrostedPhlake OX=2588135 GN=41 PE=4 SV=1
MM1 pKa = 7.46ANDD4 pKa = 4.61TIFEE8 pKa = 4.28LPEE11 pKa = 4.65IPGVTFTASYY21 pKa = 10.84GSGGEE26 pKa = 4.12TGLPSNWIRR35 pKa = 11.84IVGTVEE41 pKa = 3.86NPWYY45 pKa = 10.31DD46 pKa = 3.26PTYY49 pKa = 11.08NYY51 pKa = 11.18GLDD54 pKa = 3.77PSGHH58 pKa = 5.92TEE60 pKa = 3.79MTDD63 pKa = 2.48PWKK66 pKa = 10.72RR67 pKa = 11.84HH68 pKa = 3.86TQFPEE73 pKa = 3.62VLGPMGFEE81 pKa = 4.55GPSIGLPVDD90 pKa = 3.94PPPPPVEE97 pKa = 4.39PEE99 pKa = 3.75PTPDD103 pKa = 3.64IEE105 pKa = 4.54EE106 pKa = 4.49PTDD109 pKa = 3.42GG110 pKa = 4.69
MM1 pKa = 7.46ANDD4 pKa = 4.61TIFEE8 pKa = 4.28LPEE11 pKa = 4.65IPGVTFTASYY21 pKa = 10.84GSGGEE26 pKa = 4.12TGLPSNWIRR35 pKa = 11.84IVGTVEE41 pKa = 3.86NPWYY45 pKa = 10.31DD46 pKa = 3.26PTYY49 pKa = 11.08NYY51 pKa = 11.18GLDD54 pKa = 3.77PSGHH58 pKa = 5.92TEE60 pKa = 3.79MTDD63 pKa = 2.48PWKK66 pKa = 10.72RR67 pKa = 11.84HH68 pKa = 3.86TQFPEE73 pKa = 3.62VLGPMGFEE81 pKa = 4.55GPSIGLPVDD90 pKa = 3.94PPPPPVEE97 pKa = 4.39PEE99 pKa = 3.75PTPDD103 pKa = 3.64IEE105 pKa = 4.54EE106 pKa = 4.49PTDD109 pKa = 3.42GG110 pKa = 4.69
Molecular weight: 11.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y6EHC1|A0A4Y6EHC1_9CAUD Uncharacterized protein OS=Gordonia phage PhrostedPhlake OX=2588135 GN=53 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84SGHH5 pKa = 6.09EE6 pKa = 3.96SDD8 pKa = 3.11KK9 pKa = 10.99TRR11 pKa = 11.84ASGHH15 pKa = 4.83YY16 pKa = 8.83VVNNIVPMNNTSNTGKK32 pKa = 10.16SKK34 pKa = 10.62SVPTEE39 pKa = 3.49WQFLVGGFLRR49 pKa = 11.84HH50 pKa = 5.82LRR52 pKa = 11.84AAGQPVTTINTRR64 pKa = 11.84RR65 pKa = 11.84QQIEE69 pKa = 3.89RR70 pKa = 11.84MARR73 pKa = 11.84ALDD76 pKa = 3.84GSPAEE81 pKa = 4.29VTRR84 pKa = 11.84DD85 pKa = 3.57AITDD89 pKa = 3.16YY90 pKa = 11.03HH91 pKa = 7.2AAQTWALEE99 pKa = 3.96TRR101 pKa = 11.84RR102 pKa = 11.84GHH104 pKa = 6.45RR105 pKa = 11.84NAAVSFFAWAHH116 pKa = 5.1SCGHH120 pKa = 6.32IPGNPAQHH128 pKa = 6.15LPIVKK133 pKa = 10.05SSVPAPRR140 pKa = 11.84PAPDD144 pKa = 2.72RR145 pKa = 11.84VVNVALQTPEE155 pKa = 4.25KK156 pKa = 10.03RR157 pKa = 11.84VQLMLRR163 pKa = 11.84LSCEE167 pKa = 3.66LGLRR171 pKa = 11.84RR172 pKa = 11.84AEE174 pKa = 4.07VAVVSTTDD182 pKa = 3.16VEE184 pKa = 4.77DD185 pKa = 4.3GVGGAMLRR193 pKa = 11.84VHH195 pKa = 6.82GKK197 pKa = 9.01GGRR200 pKa = 11.84IRR202 pKa = 11.84VIPISDD208 pKa = 3.84DD209 pKa = 3.19LALRR213 pKa = 11.84IEE215 pKa = 5.0DD216 pKa = 3.65GAAGHH221 pKa = 6.29TPGAPRR227 pKa = 11.84TGYY230 pKa = 10.43LFPGNEE236 pKa = 4.26SGHH239 pKa = 6.53LSPRR243 pKa = 11.84WVGKK247 pKa = 10.12LCADD251 pKa = 4.7ALPGGWTMHH260 pKa = 5.65TLRR263 pKa = 11.84HH264 pKa = 5.69RR265 pKa = 11.84FATRR269 pKa = 11.84AYY271 pKa = 9.41RR272 pKa = 11.84GSRR275 pKa = 11.84NLRR278 pKa = 11.84AVQILLGHH286 pKa = 7.04ASLATTQRR294 pKa = 11.84YY295 pKa = 5.49TAVDD299 pKa = 3.57EE300 pKa = 4.78DD301 pKa = 4.15EE302 pKa = 4.82VRR304 pKa = 11.84AAMTSAAA311 pKa = 4.24
MM1 pKa = 7.86RR2 pKa = 11.84SGHH5 pKa = 6.09EE6 pKa = 3.96SDD8 pKa = 3.11KK9 pKa = 10.99TRR11 pKa = 11.84ASGHH15 pKa = 4.83YY16 pKa = 8.83VVNNIVPMNNTSNTGKK32 pKa = 10.16SKK34 pKa = 10.62SVPTEE39 pKa = 3.49WQFLVGGFLRR49 pKa = 11.84HH50 pKa = 5.82LRR52 pKa = 11.84AAGQPVTTINTRR64 pKa = 11.84RR65 pKa = 11.84QQIEE69 pKa = 3.89RR70 pKa = 11.84MARR73 pKa = 11.84ALDD76 pKa = 3.84GSPAEE81 pKa = 4.29VTRR84 pKa = 11.84DD85 pKa = 3.57AITDD89 pKa = 3.16YY90 pKa = 11.03HH91 pKa = 7.2AAQTWALEE99 pKa = 3.96TRR101 pKa = 11.84RR102 pKa = 11.84GHH104 pKa = 6.45RR105 pKa = 11.84NAAVSFFAWAHH116 pKa = 5.1SCGHH120 pKa = 6.32IPGNPAQHH128 pKa = 6.15LPIVKK133 pKa = 10.05SSVPAPRR140 pKa = 11.84PAPDD144 pKa = 2.72RR145 pKa = 11.84VVNVALQTPEE155 pKa = 4.25KK156 pKa = 10.03RR157 pKa = 11.84VQLMLRR163 pKa = 11.84LSCEE167 pKa = 3.66LGLRR171 pKa = 11.84RR172 pKa = 11.84AEE174 pKa = 4.07VAVVSTTDD182 pKa = 3.16VEE184 pKa = 4.77DD185 pKa = 4.3GVGGAMLRR193 pKa = 11.84VHH195 pKa = 6.82GKK197 pKa = 9.01GGRR200 pKa = 11.84IRR202 pKa = 11.84VIPISDD208 pKa = 3.84DD209 pKa = 3.19LALRR213 pKa = 11.84IEE215 pKa = 5.0DD216 pKa = 3.65GAAGHH221 pKa = 6.29TPGAPRR227 pKa = 11.84TGYY230 pKa = 10.43LFPGNEE236 pKa = 4.26SGHH239 pKa = 6.53LSPRR243 pKa = 11.84WVGKK247 pKa = 10.12LCADD251 pKa = 4.7ALPGGWTMHH260 pKa = 5.65TLRR263 pKa = 11.84HH264 pKa = 5.69RR265 pKa = 11.84FATRR269 pKa = 11.84AYY271 pKa = 9.41RR272 pKa = 11.84GSRR275 pKa = 11.84NLRR278 pKa = 11.84AVQILLGHH286 pKa = 7.04ASLATTQRR294 pKa = 11.84YY295 pKa = 5.49TAVDD299 pKa = 3.57EE300 pKa = 4.78DD301 pKa = 4.15EE302 pKa = 4.82VRR304 pKa = 11.84AAMTSAAA311 pKa = 4.24
Molecular weight: 33.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
16976 |
53 |
1846 |
192.9 |
21.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.628 ± 0.539 | 1.031 ± 0.18 |
6.916 ± 0.319 | 5.578 ± 0.233 |
2.763 ± 0.125 | 8.683 ± 0.307 |
2.203 ± 0.159 | 4.418 ± 0.17 |
3.187 ± 0.235 | 7.458 ± 0.252 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.233 ± 0.163 | 2.945 ± 0.139 |
6.05 ± 0.282 | 3.358 ± 0.126 |
7.522 ± 0.32 | 5.219 ± 0.158 |
6.68 ± 0.239 | 7.782 ± 0.221 |
2.062 ± 0.119 | 2.286 ± 0.17 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
