
Thalassocella blandensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Thalassocella
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
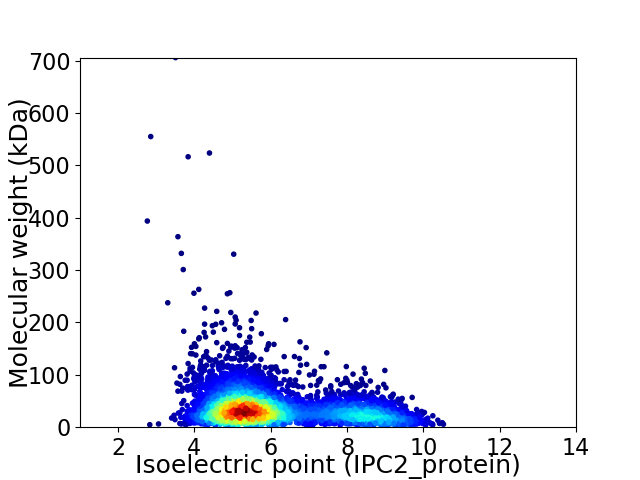
Virtual 2D-PAGE plot for 4758 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A509E4S0|A0A509E4S0_9RHIZ Cation efflux system protein CusA OS=Thalassocella blandensis OX=2584524 GN=cusA PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.6LLRR5 pKa = 11.84TTLGMLSASLVLVACGGGGGGGGGGGGSTPTPTPDD40 pKa = 4.1PGVSCNDD47 pKa = 3.28GTPSFVTCTDD57 pKa = 3.37GQNNGPDD64 pKa = 3.64VYY66 pKa = 9.68TLHH69 pKa = 6.18GTVDD73 pKa = 3.39ADD75 pKa = 3.52YY76 pKa = 11.08TMDD79 pKa = 4.73SSYY82 pKa = 10.68EE83 pKa = 3.66WRR85 pKa = 11.84LDD87 pKa = 3.32GVVKK91 pKa = 10.73VGSGNVEE98 pKa = 3.81VTSADD103 pKa = 3.38DD104 pKa = 3.26VAAIQDD110 pKa = 3.7AGVTLTIEE118 pKa = 4.54PGTHH122 pKa = 5.69IKK124 pKa = 10.93AFDD127 pKa = 4.22DD128 pKa = 4.3GVLLVTRR135 pKa = 11.84GSKK138 pKa = 10.49LMADD142 pKa = 4.54GTATAPITFSSLDD155 pKa = 3.62DD156 pKa = 4.79GYY158 pKa = 11.44DD159 pKa = 4.34GIGEE163 pKa = 4.07WGGVIVQGFAPQYY176 pKa = 9.18GQGDD180 pKa = 3.76TGPCYY185 pKa = 10.08GTGSICNVEE194 pKa = 4.25GEE196 pKa = 4.56GGTEE200 pKa = 3.83VAVYY204 pKa = 10.42GGDD207 pKa = 3.88IEE209 pKa = 5.57DD210 pKa = 5.01DD211 pKa = 3.38NSGVIRR217 pKa = 11.84YY218 pKa = 8.28VRR220 pKa = 11.84IAEE223 pKa = 4.41GGLIAGPNNEE233 pKa = 4.28INGLTLQGVGHH244 pKa = 6.05GTVVDD249 pKa = 4.06YY250 pKa = 10.97VHH252 pKa = 6.4VHH254 pKa = 6.05GNLDD258 pKa = 4.89DD259 pKa = 4.22GVEE262 pKa = 4.17WFGGTVNVTHH272 pKa = 7.34LVLTNNDD279 pKa = 3.85DD280 pKa = 4.09DD281 pKa = 7.11DD282 pKa = 5.38IDD284 pKa = 4.01FDD286 pKa = 4.14EE287 pKa = 5.78GYY289 pKa = 10.36KK290 pKa = 11.14GNIQYY295 pKa = 10.91AIVVKK300 pKa = 10.36DD301 pKa = 3.53QTKK304 pKa = 9.61EE305 pKa = 3.89APTGSNDD312 pKa = 3.2PRR314 pKa = 11.84GIEE317 pKa = 4.38ANSSDD322 pKa = 4.53DD323 pKa = 5.44DD324 pKa = 4.19YY325 pKa = 12.22VPDD328 pKa = 4.07TNAVLANVTVIGGAVNNAEE347 pKa = 4.32GKK349 pKa = 9.7EE350 pKa = 4.03EE351 pKa = 4.07PGMRR355 pKa = 11.84LRR357 pKa = 11.84GSLTATIYY365 pKa = 10.83NSAVKK370 pKa = 10.54GFDD373 pKa = 3.29TGCIRR378 pKa = 11.84IDD380 pKa = 4.86DD381 pKa = 4.44SDD383 pKa = 3.9TDD385 pKa = 4.15GDD387 pKa = 3.94GTADD391 pKa = 4.38DD392 pKa = 4.73FSDD395 pKa = 3.64VNLINVFGDD404 pKa = 3.96CADD407 pKa = 3.33GFYY410 pKa = 10.99DD411 pKa = 6.01KK412 pKa = 10.83RR413 pKa = 11.84AADD416 pKa = 3.61NASNAVVSSVTLDD429 pKa = 2.99SAYY432 pKa = 10.83ALTEE436 pKa = 3.95SAATLGAAPAINAIVNGSGFMFDD459 pKa = 2.94QTAYY463 pKa = 10.18VGAVAPGTAMEE474 pKa = 4.45DD475 pKa = 3.22AWWYY479 pKa = 9.63GWTIPGVLSDD489 pKa = 5.45AEE491 pKa = 4.66AGDD494 pKa = 4.28DD495 pKa = 4.17APAEE499 pKa = 4.05ADD501 pKa = 3.79FVNCNHH507 pKa = 7.12ALLEE511 pKa = 4.3CTVAGTIDD519 pKa = 3.53EE520 pKa = 5.52DD521 pKa = 4.25YY522 pKa = 10.3TFVSGYY528 pKa = 7.49TWFLDD533 pKa = 3.49GTVKK537 pKa = 10.66VGNGNIEE544 pKa = 4.17VASDD548 pKa = 3.41ADD550 pKa = 3.65VAAIQAAGVTLTVQAGTNVKK570 pKa = 10.59AFDD573 pKa = 4.94DD574 pKa = 4.04GTLLVTRR581 pKa = 11.84GSKK584 pKa = 10.49LMADD588 pKa = 4.4GTAAAPITFSSLDD601 pKa = 3.41QNFSSEE607 pKa = 4.43GEE609 pKa = 3.87WGGLIIQGFAPQYY622 pKa = 9.47GQGDD626 pKa = 3.7TGPCYY631 pKa = 10.92GEE633 pKa = 3.86GTTCNVAGEE642 pKa = 4.68GGTDD646 pKa = 3.29VAVYY650 pKa = 10.13GGNIVDD656 pKa = 4.83DD657 pKa = 3.98NSGVIRR663 pKa = 11.84YY664 pKa = 8.28VRR666 pKa = 11.84IAEE669 pKa = 4.45GGLVAGPNNEE679 pKa = 4.04INGLTLQGVGYY690 pKa = 7.82GTTIEE695 pKa = 4.15YY696 pKa = 10.15VQVHH700 pKa = 6.39GNLDD704 pKa = 4.07DD705 pKa = 4.18GVEE708 pKa = 4.17WFGGTVNAKK717 pKa = 10.38YY718 pKa = 10.37LVLTNNDD725 pKa = 3.83DD726 pKa = 4.09DD727 pKa = 7.11DD728 pKa = 5.38IDD730 pKa = 4.01FDD732 pKa = 4.14EE733 pKa = 5.78GYY735 pKa = 10.36KK736 pKa = 11.14GNIQYY741 pKa = 10.94AIVVKK746 pKa = 10.02NQNKK750 pKa = 9.41AVPTGSNDD758 pKa = 3.1PRR760 pKa = 11.84GIEE763 pKa = 4.38ANSSDD768 pKa = 4.82DD769 pKa = 5.22DD770 pKa = 4.26FVPDD774 pKa = 4.11TNAVLANITLIGGPVNNSTDD794 pKa = 3.39KK795 pKa = 11.03EE796 pKa = 4.38EE797 pKa = 4.49PGMRR801 pKa = 11.84LRR803 pKa = 11.84GSLTVAIHH811 pKa = 4.87NTALKK816 pKa = 10.74GFDD819 pKa = 3.6TGCVRR824 pKa = 11.84VDD826 pKa = 4.9DD827 pKa = 4.89SDD829 pKa = 3.81TDD831 pKa = 4.04GDD833 pKa = 3.94GTADD837 pKa = 3.72DD838 pKa = 4.76FSNVTLNNIIADD850 pKa = 4.11CAGGFYY856 pKa = 10.79AKK858 pKa = 10.86GEE860 pKa = 4.08ADD862 pKa = 3.63TVNNAIEE869 pKa = 4.32STVTIDD875 pKa = 3.62EE876 pKa = 4.55AFALTDD882 pKa = 3.46AAASLGVTTAITAMDD897 pKa = 4.53NGSMFAFDD905 pKa = 3.18QTSFVGAVAPGTAAAEE921 pKa = 4.14AWWAGWTLEE930 pKa = 4.12GTLDD934 pKa = 4.91DD935 pKa = 5.91AIEE938 pKa = 4.13NNN940 pKa = 3.7
MM1 pKa = 7.49KK2 pKa = 10.6LLRR5 pKa = 11.84TTLGMLSASLVLVACGGGGGGGGGGGGSTPTPTPDD40 pKa = 4.1PGVSCNDD47 pKa = 3.28GTPSFVTCTDD57 pKa = 3.37GQNNGPDD64 pKa = 3.64VYY66 pKa = 9.68TLHH69 pKa = 6.18GTVDD73 pKa = 3.39ADD75 pKa = 3.52YY76 pKa = 11.08TMDD79 pKa = 4.73SSYY82 pKa = 10.68EE83 pKa = 3.66WRR85 pKa = 11.84LDD87 pKa = 3.32GVVKK91 pKa = 10.73VGSGNVEE98 pKa = 3.81VTSADD103 pKa = 3.38DD104 pKa = 3.26VAAIQDD110 pKa = 3.7AGVTLTIEE118 pKa = 4.54PGTHH122 pKa = 5.69IKK124 pKa = 10.93AFDD127 pKa = 4.22DD128 pKa = 4.3GVLLVTRR135 pKa = 11.84GSKK138 pKa = 10.49LMADD142 pKa = 4.54GTATAPITFSSLDD155 pKa = 3.62DD156 pKa = 4.79GYY158 pKa = 11.44DD159 pKa = 4.34GIGEE163 pKa = 4.07WGGVIVQGFAPQYY176 pKa = 9.18GQGDD180 pKa = 3.76TGPCYY185 pKa = 10.08GTGSICNVEE194 pKa = 4.25GEE196 pKa = 4.56GGTEE200 pKa = 3.83VAVYY204 pKa = 10.42GGDD207 pKa = 3.88IEE209 pKa = 5.57DD210 pKa = 5.01DD211 pKa = 3.38NSGVIRR217 pKa = 11.84YY218 pKa = 8.28VRR220 pKa = 11.84IAEE223 pKa = 4.41GGLIAGPNNEE233 pKa = 4.28INGLTLQGVGHH244 pKa = 6.05GTVVDD249 pKa = 4.06YY250 pKa = 10.97VHH252 pKa = 6.4VHH254 pKa = 6.05GNLDD258 pKa = 4.89DD259 pKa = 4.22GVEE262 pKa = 4.17WFGGTVNVTHH272 pKa = 7.34LVLTNNDD279 pKa = 3.85DD280 pKa = 4.09DD281 pKa = 7.11DD282 pKa = 5.38IDD284 pKa = 4.01FDD286 pKa = 4.14EE287 pKa = 5.78GYY289 pKa = 10.36KK290 pKa = 11.14GNIQYY295 pKa = 10.91AIVVKK300 pKa = 10.36DD301 pKa = 3.53QTKK304 pKa = 9.61EE305 pKa = 3.89APTGSNDD312 pKa = 3.2PRR314 pKa = 11.84GIEE317 pKa = 4.38ANSSDD322 pKa = 4.53DD323 pKa = 5.44DD324 pKa = 4.19YY325 pKa = 12.22VPDD328 pKa = 4.07TNAVLANVTVIGGAVNNAEE347 pKa = 4.32GKK349 pKa = 9.7EE350 pKa = 4.03EE351 pKa = 4.07PGMRR355 pKa = 11.84LRR357 pKa = 11.84GSLTATIYY365 pKa = 10.83NSAVKK370 pKa = 10.54GFDD373 pKa = 3.29TGCIRR378 pKa = 11.84IDD380 pKa = 4.86DD381 pKa = 4.44SDD383 pKa = 3.9TDD385 pKa = 4.15GDD387 pKa = 3.94GTADD391 pKa = 4.38DD392 pKa = 4.73FSDD395 pKa = 3.64VNLINVFGDD404 pKa = 3.96CADD407 pKa = 3.33GFYY410 pKa = 10.99DD411 pKa = 6.01KK412 pKa = 10.83RR413 pKa = 11.84AADD416 pKa = 3.61NASNAVVSSVTLDD429 pKa = 2.99SAYY432 pKa = 10.83ALTEE436 pKa = 3.95SAATLGAAPAINAIVNGSGFMFDD459 pKa = 2.94QTAYY463 pKa = 10.18VGAVAPGTAMEE474 pKa = 4.45DD475 pKa = 3.22AWWYY479 pKa = 9.63GWTIPGVLSDD489 pKa = 5.45AEE491 pKa = 4.66AGDD494 pKa = 4.28DD495 pKa = 4.17APAEE499 pKa = 4.05ADD501 pKa = 3.79FVNCNHH507 pKa = 7.12ALLEE511 pKa = 4.3CTVAGTIDD519 pKa = 3.53EE520 pKa = 5.52DD521 pKa = 4.25YY522 pKa = 10.3TFVSGYY528 pKa = 7.49TWFLDD533 pKa = 3.49GTVKK537 pKa = 10.66VGNGNIEE544 pKa = 4.17VASDD548 pKa = 3.41ADD550 pKa = 3.65VAAIQAAGVTLTVQAGTNVKK570 pKa = 10.59AFDD573 pKa = 4.94DD574 pKa = 4.04GTLLVTRR581 pKa = 11.84GSKK584 pKa = 10.49LMADD588 pKa = 4.4GTAAAPITFSSLDD601 pKa = 3.41QNFSSEE607 pKa = 4.43GEE609 pKa = 3.87WGGLIIQGFAPQYY622 pKa = 9.47GQGDD626 pKa = 3.7TGPCYY631 pKa = 10.92GEE633 pKa = 3.86GTTCNVAGEE642 pKa = 4.68GGTDD646 pKa = 3.29VAVYY650 pKa = 10.13GGNIVDD656 pKa = 4.83DD657 pKa = 3.98NSGVIRR663 pKa = 11.84YY664 pKa = 8.28VRR666 pKa = 11.84IAEE669 pKa = 4.45GGLVAGPNNEE679 pKa = 4.04INGLTLQGVGYY690 pKa = 7.82GTTIEE695 pKa = 4.15YY696 pKa = 10.15VQVHH700 pKa = 6.39GNLDD704 pKa = 4.07DD705 pKa = 4.18GVEE708 pKa = 4.17WFGGTVNAKK717 pKa = 10.38YY718 pKa = 10.37LVLTNNDD725 pKa = 3.83DD726 pKa = 4.09DD727 pKa = 7.11DD728 pKa = 5.38IDD730 pKa = 4.01FDD732 pKa = 4.14EE733 pKa = 5.78GYY735 pKa = 10.36KK736 pKa = 11.14GNIQYY741 pKa = 10.94AIVVKK746 pKa = 10.02NQNKK750 pKa = 9.41AVPTGSNDD758 pKa = 3.1PRR760 pKa = 11.84GIEE763 pKa = 4.38ANSSDD768 pKa = 4.82DD769 pKa = 5.22DD770 pKa = 4.26FVPDD774 pKa = 4.11TNAVLANITLIGGPVNNSTDD794 pKa = 3.39KK795 pKa = 11.03EE796 pKa = 4.38EE797 pKa = 4.49PGMRR801 pKa = 11.84LRR803 pKa = 11.84GSLTVAIHH811 pKa = 4.87NTALKK816 pKa = 10.74GFDD819 pKa = 3.6TGCVRR824 pKa = 11.84VDD826 pKa = 4.9DD827 pKa = 4.89SDD829 pKa = 3.81TDD831 pKa = 4.04GDD833 pKa = 3.94GTADD837 pKa = 3.72DD838 pKa = 4.76FSNVTLNNIIADD850 pKa = 4.11CAGGFYY856 pKa = 10.79AKK858 pKa = 10.86GEE860 pKa = 4.08ADD862 pKa = 3.63TVNNAIEE869 pKa = 4.32STVTIDD875 pKa = 3.62EE876 pKa = 4.55AFALTDD882 pKa = 3.46AAASLGVTTAITAMDD897 pKa = 4.53NGSMFAFDD905 pKa = 3.18QTSFVGAVAPGTAAAEE921 pKa = 4.14AWWAGWTLEE930 pKa = 4.12GTLDD934 pKa = 4.91DD935 pKa = 5.91AIEE938 pKa = 4.13NNN940 pKa = 3.7
Molecular weight: 96.71 kDa
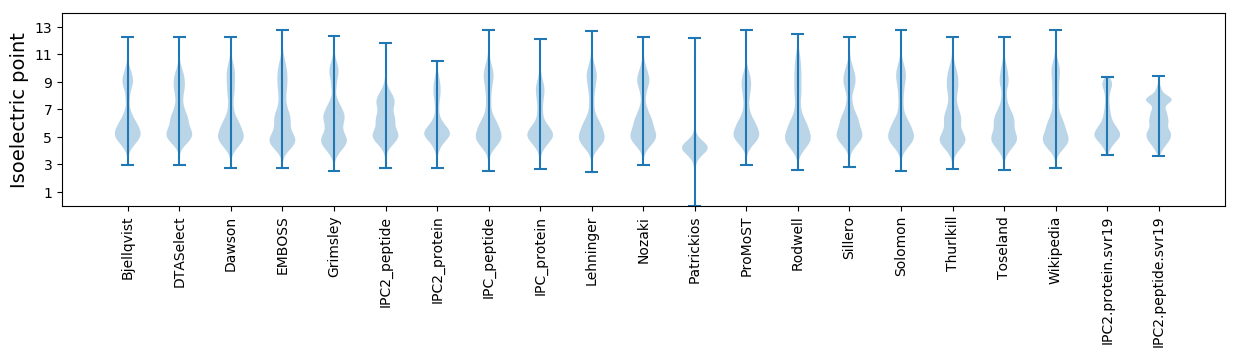
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A509E2X4|A0A509E2X4_9RHIZ Beta-glucanase OS=Thalassocella blandensis OX=2584524 GN=bglA_9 PE=4 SV=1
MM1 pKa = 7.34VRR3 pKa = 11.84LPEE6 pKa = 4.06NKK8 pKa = 9.31IPAAKK13 pKa = 10.17FIIARR18 pKa = 11.84KK19 pKa = 9.02RR20 pKa = 11.84MRR22 pKa = 11.84IEE24 pKa = 4.09GFTIGAKK31 pKa = 7.38TACTMALWCYY41 pKa = 9.59FAAIPSQYY49 pKa = 9.74FSRR52 pKa = 11.84IRR54 pKa = 11.84RR55 pKa = 11.84FQMLNDD61 pKa = 3.6VKK63 pKa = 11.17
MM1 pKa = 7.34VRR3 pKa = 11.84LPEE6 pKa = 4.06NKK8 pKa = 9.31IPAAKK13 pKa = 10.17FIIARR18 pKa = 11.84KK19 pKa = 9.02RR20 pKa = 11.84MRR22 pKa = 11.84IEE24 pKa = 4.09GFTIGAKK31 pKa = 7.38TACTMALWCYY41 pKa = 9.59FAAIPSQYY49 pKa = 9.74FSRR52 pKa = 11.84IRR54 pKa = 11.84RR55 pKa = 11.84FQMLNDD61 pKa = 3.6VKK63 pKa = 11.17
Molecular weight: 7.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1693339 |
29 |
6820 |
355.9 |
39.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.536 ± 0.041 | 1.033 ± 0.013 |
5.81 ± 0.04 | 6.344 ± 0.039 |
4.258 ± 0.023 | 6.919 ± 0.046 |
2.236 ± 0.022 | 6.222 ± 0.029 |
5.064 ± 0.047 | 9.695 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.284 ± 0.019 | 4.673 ± 0.037 |
3.933 ± 0.025 | 4.409 ± 0.03 |
4.571 ± 0.032 | 7.278 ± 0.041 |
5.483 ± 0.043 | 6.782 ± 0.036 |
1.3 ± 0.017 | 3.17 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
