
Methanopyrus kandleri (strain AV19 / DSM 6324 / JCM 9639 / NBRC 100938)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanopyri; Methanopyrales; Methanopyraceae; Methanopyrus; Methanopyrus kandleri
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
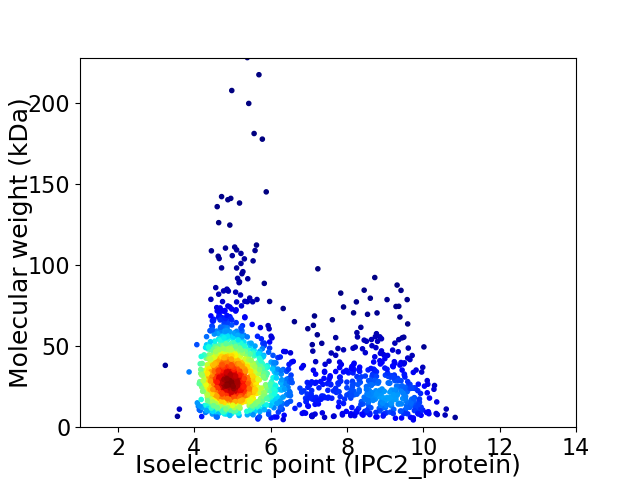
Virtual 2D-PAGE plot for 1687 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8TGZ6|Q8TGZ6_METKA tRNA/rRNA cytosine-C5-methylase OS=Methanopyrus kandleri (strain AV19 / DSM 6324 / JCM 9639 / NBRC 100938) OX=190192 GN=MK0370 PE=3 SV=1
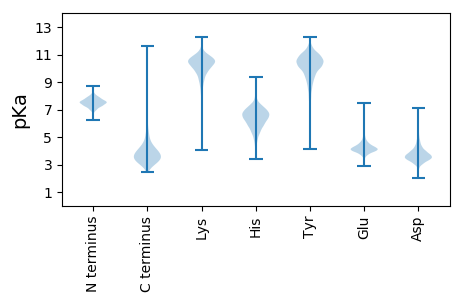
MM1 pKa = 7.53VSVNEE6 pKa = 4.03NALPLVEE13 pKa = 5.06RR14 pKa = 11.84MIEE17 pKa = 3.96RR18 pKa = 11.84AEE20 pKa = 3.93LLNVEE25 pKa = 4.32VQEE28 pKa = 4.59LEE30 pKa = 4.0NGTTVIDD37 pKa = 4.41CGVEE41 pKa = 3.57AAGGFEE47 pKa = 4.07AGLLFSEE54 pKa = 4.79VCMGGLATVEE64 pKa = 4.03LTEE67 pKa = 4.5FEE69 pKa = 4.6HH70 pKa = 7.9DD71 pKa = 4.26GLCLPAVQVTTDD83 pKa = 3.37HH84 pKa = 6.91PAVSTLAAQKK94 pKa = 10.59AGWQVQVGDD103 pKa = 4.04YY104 pKa = 10.55FAMGSGPARR113 pKa = 11.84ALALKK118 pKa = 10.2PKK120 pKa = 8.62EE121 pKa = 4.2TYY123 pKa = 10.53EE124 pKa = 4.26EE125 pKa = 3.73IDD127 pKa = 3.77YY128 pKa = 11.47EE129 pKa = 5.04DD130 pKa = 4.72DD131 pKa = 3.69ADD133 pKa = 4.28VAILCLEE140 pKa = 4.18SSEE143 pKa = 5.83LPDD146 pKa = 3.38EE147 pKa = 5.07DD148 pKa = 3.93VAEE151 pKa = 4.56HH152 pKa = 6.34VADD155 pKa = 3.8EE156 pKa = 4.89CGVDD160 pKa = 3.87PEE162 pKa = 4.17NLYY165 pKa = 11.32LLVAPTASIVGSVQVSARR183 pKa = 11.84VVEE186 pKa = 4.15TGLYY190 pKa = 10.04KK191 pKa = 10.75LLEE194 pKa = 4.06VLEE197 pKa = 4.34YY198 pKa = 11.0DD199 pKa = 3.79VTRR202 pKa = 11.84VKK204 pKa = 10.79YY205 pKa = 9.17ATGTAPIAPVADD217 pKa = 4.38DD218 pKa = 4.93DD219 pKa = 4.6GEE221 pKa = 4.97AMGRR225 pKa = 11.84TNDD228 pKa = 3.9CILYY232 pKa = 10.24GGTVYY237 pKa = 10.79LYY239 pKa = 11.15VEE241 pKa = 4.98GDD243 pKa = 3.65DD244 pKa = 4.06EE245 pKa = 4.66LPEE248 pKa = 4.18VVEE251 pKa = 4.4EE252 pKa = 4.52LPSEE256 pKa = 4.1ASEE259 pKa = 5.43DD260 pKa = 3.61YY261 pKa = 10.72GKK263 pKa = 9.98PFMKK267 pKa = 9.89IFEE270 pKa = 4.14EE271 pKa = 4.34ADD273 pKa = 3.08YY274 pKa = 11.18DD275 pKa = 4.33FYY277 pKa = 11.55KK278 pKa = 10.22IDD280 pKa = 4.08PGVFAPARR288 pKa = 11.84VVVNDD293 pKa = 4.07LSTGKK298 pKa = 8.8TYY300 pKa = 9.51TAGEE304 pKa = 4.21INVDD308 pKa = 3.57VLKK311 pKa = 10.92EE312 pKa = 4.18SFGLL316 pKa = 3.77
MM1 pKa = 7.53VSVNEE6 pKa = 4.03NALPLVEE13 pKa = 5.06RR14 pKa = 11.84MIEE17 pKa = 3.96RR18 pKa = 11.84AEE20 pKa = 3.93LLNVEE25 pKa = 4.32VQEE28 pKa = 4.59LEE30 pKa = 4.0NGTTVIDD37 pKa = 4.41CGVEE41 pKa = 3.57AAGGFEE47 pKa = 4.07AGLLFSEE54 pKa = 4.79VCMGGLATVEE64 pKa = 4.03LTEE67 pKa = 4.5FEE69 pKa = 4.6HH70 pKa = 7.9DD71 pKa = 4.26GLCLPAVQVTTDD83 pKa = 3.37HH84 pKa = 6.91PAVSTLAAQKK94 pKa = 10.59AGWQVQVGDD103 pKa = 4.04YY104 pKa = 10.55FAMGSGPARR113 pKa = 11.84ALALKK118 pKa = 10.2PKK120 pKa = 8.62EE121 pKa = 4.2TYY123 pKa = 10.53EE124 pKa = 4.26EE125 pKa = 3.73IDD127 pKa = 3.77YY128 pKa = 11.47EE129 pKa = 5.04DD130 pKa = 4.72DD131 pKa = 3.69ADD133 pKa = 4.28VAILCLEE140 pKa = 4.18SSEE143 pKa = 5.83LPDD146 pKa = 3.38EE147 pKa = 5.07DD148 pKa = 3.93VAEE151 pKa = 4.56HH152 pKa = 6.34VADD155 pKa = 3.8EE156 pKa = 4.89CGVDD160 pKa = 3.87PEE162 pKa = 4.17NLYY165 pKa = 11.32LLVAPTASIVGSVQVSARR183 pKa = 11.84VVEE186 pKa = 4.15TGLYY190 pKa = 10.04KK191 pKa = 10.75LLEE194 pKa = 4.06VLEE197 pKa = 4.34YY198 pKa = 11.0DD199 pKa = 3.79VTRR202 pKa = 11.84VKK204 pKa = 10.79YY205 pKa = 9.17ATGTAPIAPVADD217 pKa = 4.38DD218 pKa = 4.93DD219 pKa = 4.6GEE221 pKa = 4.97AMGRR225 pKa = 11.84TNDD228 pKa = 3.9CILYY232 pKa = 10.24GGTVYY237 pKa = 10.79LYY239 pKa = 11.15VEE241 pKa = 4.98GDD243 pKa = 3.65DD244 pKa = 4.06EE245 pKa = 4.66LPEE248 pKa = 4.18VVEE251 pKa = 4.4EE252 pKa = 4.52LPSEE256 pKa = 4.1ASEE259 pKa = 5.43DD260 pKa = 3.61YY261 pKa = 10.72GKK263 pKa = 9.98PFMKK267 pKa = 9.89IFEE270 pKa = 4.14EE271 pKa = 4.34ADD273 pKa = 3.08YY274 pKa = 11.18DD275 pKa = 4.33FYY277 pKa = 11.55KK278 pKa = 10.22IDD280 pKa = 4.08PGVFAPARR288 pKa = 11.84VVVNDD293 pKa = 4.07LSTGKK298 pKa = 8.8TYY300 pKa = 9.51TAGEE304 pKa = 4.21INVDD308 pKa = 3.57VLKK311 pKa = 10.92EE312 pKa = 4.18SFGLL316 pKa = 3.77
Molecular weight: 34.04 kDa
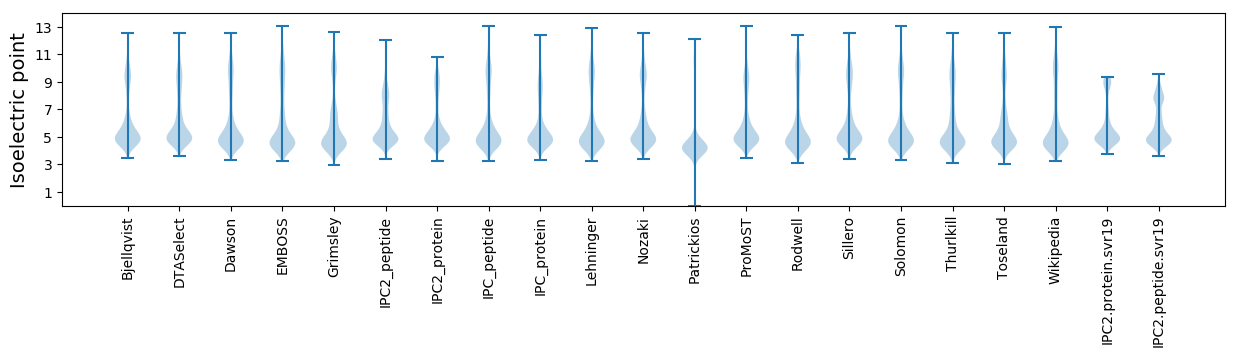
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8TUY4|RL31_METKA 50S ribosomal protein L31e OS=Methanopyrus kandleri (strain AV19 / DSM 6324 / JCM 9639 / NBRC 100938) OX=190192 GN=rpl31e PE=3 SV=1
MM1 pKa = 7.51ARR3 pKa = 11.84VKK5 pKa = 10.61PLGKK9 pKa = 9.72KK10 pKa = 10.03LRR12 pKa = 11.84MAKK15 pKa = 10.06AIKK18 pKa = 9.32QNRR21 pKa = 11.84RR22 pKa = 11.84VPPWVVAKK30 pKa = 10.08TGGRR34 pKa = 11.84VIDD37 pKa = 3.78NPKK40 pKa = 8.35RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.1WRR45 pKa = 11.84RR46 pKa = 11.84SKK48 pKa = 10.9LKK50 pKa = 9.79PP51 pKa = 3.26
MM1 pKa = 7.51ARR3 pKa = 11.84VKK5 pKa = 10.61PLGKK9 pKa = 9.72KK10 pKa = 10.03LRR12 pKa = 11.84MAKK15 pKa = 10.06AIKK18 pKa = 9.32QNRR21 pKa = 11.84RR22 pKa = 11.84VPPWVVAKK30 pKa = 10.08TGGRR34 pKa = 11.84VIDD37 pKa = 3.78NPKK40 pKa = 8.35RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.1WRR45 pKa = 11.84RR46 pKa = 11.84SKK48 pKa = 10.9LKK50 pKa = 9.79PP51 pKa = 3.26
Molecular weight: 6.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
500958 |
39 |
2042 |
297.0 |
33.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.343 ± 0.064 | 1.32 ± 0.029 |
5.798 ± 0.048 | 10.0 ± 0.099 |
2.873 ± 0.031 | 8.051 ± 0.059 |
1.931 ± 0.022 | 4.827 ± 0.051 |
4.022 ± 0.056 | 10.084 ± 0.081 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.9 ± 0.027 | 1.916 ± 0.027 |
5.463 ± 0.045 | 1.409 ± 0.028 |
8.345 ± 0.076 | 4.621 ± 0.05 |
4.594 ± 0.041 | 10.442 ± 0.06 |
1.239 ± 0.029 | 2.821 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
