
Aureimonas altamirensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomic
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
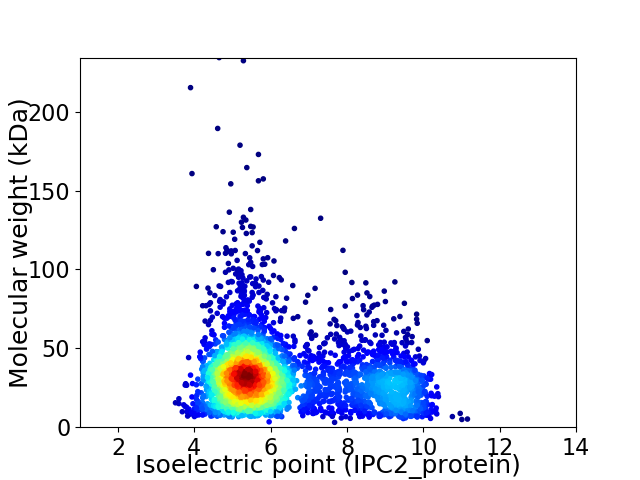
Virtual 2D-PAGE plot for 3519 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
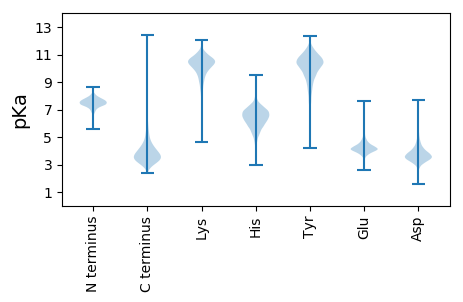
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B1Q2E7|A0A0B1Q2E7_9RHIZ Uncharacterized protein OS=Aureimonas altamirensis OX=370622 GN=LA66_10610 PE=4 SV=1
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALVAVSGARR21 pKa = 11.84AADD24 pKa = 4.4AIVMVEE30 pKa = 4.17PEE32 pKa = 3.81PVEE35 pKa = 3.97YY36 pKa = 11.07VRR38 pKa = 11.84VCDD41 pKa = 3.81VYY43 pKa = 10.81GTGFYY48 pKa = 10.01YY49 pKa = 9.98IPGTEE54 pKa = 4.14TCIKK58 pKa = 8.83TSGYY62 pKa = 10.83LRR64 pKa = 11.84FQYY67 pKa = 10.64NVAGSPAGNSEE78 pKa = 3.64GDD80 pKa = 3.67DD81 pKa = 3.8YY82 pKa = 11.79QALSSVRR89 pKa = 11.84ARR91 pKa = 11.84LNLDD95 pKa = 2.98VRR97 pKa = 11.84EE98 pKa = 4.01EE99 pKa = 4.52TEE101 pKa = 4.23LGTLRR106 pKa = 11.84AYY108 pKa = 10.36FRR110 pKa = 11.84LQAQNTGASPEE121 pKa = 4.53DD122 pKa = 3.94EE123 pKa = 4.21FAMEE127 pKa = 4.25QGYY130 pKa = 8.6LQLGGFLAGYY140 pKa = 10.22RR141 pKa = 11.84DD142 pKa = 4.32TLWSSDD148 pKa = 2.76IGGIEE153 pKa = 4.96DD154 pKa = 5.08GLPTDD159 pKa = 4.89DD160 pKa = 5.75DD161 pKa = 4.69LPIGDD166 pKa = 5.05FNTNQISYY174 pKa = 8.46TFATGGFAATLGLEE188 pKa = 4.38DD189 pKa = 5.96DD190 pKa = 4.38GTGDD194 pKa = 3.63IAPDD198 pKa = 3.34VHH200 pKa = 6.86GKK202 pKa = 10.3LSYY205 pKa = 8.91TGAWGGVFLSAVYY218 pKa = 10.68DD219 pKa = 3.59EE220 pKa = 4.89TFNAEE225 pKa = 4.16DD226 pKa = 3.5VTDD229 pKa = 4.91LFGIDD234 pKa = 5.57DD235 pKa = 4.92LDD237 pKa = 4.37LPAFFPFIPEE247 pKa = 3.84RR248 pKa = 11.84DD249 pKa = 3.3VDD251 pKa = 3.87GSNDD255 pKa = 3.23AFALKK260 pKa = 10.53AGLTLQDD267 pKa = 4.62LLVADD272 pKa = 4.2STLKK276 pKa = 10.71VDD278 pKa = 3.81GHH280 pKa = 6.08WASDD284 pKa = 3.44PTSYY288 pKa = 11.4AVISGFTSAGGILPSNATLEE308 pKa = 4.26NPTPGAFPLVLEE320 pKa = 4.45WSVGAGYY327 pKa = 8.38SQKK330 pKa = 10.28FNRR333 pKa = 11.84VTAGVSGRR341 pKa = 11.84YY342 pKa = 9.77GEE344 pKa = 4.26TFDD347 pKa = 3.88YY348 pKa = 11.27AFVAPEE354 pKa = 3.99VPGGFVFSSADD365 pKa = 3.19YY366 pKa = 8.63WAVVGDD372 pKa = 3.74IGYY375 pKa = 10.2DD376 pKa = 3.1LTANMNVLAEE386 pKa = 4.17VKK388 pKa = 10.08YY389 pKa = 10.88QNIDD393 pKa = 3.37FGGVASDD400 pKa = 4.04DD401 pKa = 3.68QTSGFLRR408 pKa = 11.84FQRR411 pKa = 11.84NFF413 pKa = 3.06
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALVAVSGARR21 pKa = 11.84AADD24 pKa = 4.4AIVMVEE30 pKa = 4.17PEE32 pKa = 3.81PVEE35 pKa = 3.97YY36 pKa = 11.07VRR38 pKa = 11.84VCDD41 pKa = 3.81VYY43 pKa = 10.81GTGFYY48 pKa = 10.01YY49 pKa = 9.98IPGTEE54 pKa = 4.14TCIKK58 pKa = 8.83TSGYY62 pKa = 10.83LRR64 pKa = 11.84FQYY67 pKa = 10.64NVAGSPAGNSEE78 pKa = 3.64GDD80 pKa = 3.67DD81 pKa = 3.8YY82 pKa = 11.79QALSSVRR89 pKa = 11.84ARR91 pKa = 11.84LNLDD95 pKa = 2.98VRR97 pKa = 11.84EE98 pKa = 4.01EE99 pKa = 4.52TEE101 pKa = 4.23LGTLRR106 pKa = 11.84AYY108 pKa = 10.36FRR110 pKa = 11.84LQAQNTGASPEE121 pKa = 4.53DD122 pKa = 3.94EE123 pKa = 4.21FAMEE127 pKa = 4.25QGYY130 pKa = 8.6LQLGGFLAGYY140 pKa = 10.22RR141 pKa = 11.84DD142 pKa = 4.32TLWSSDD148 pKa = 2.76IGGIEE153 pKa = 4.96DD154 pKa = 5.08GLPTDD159 pKa = 4.89DD160 pKa = 5.75DD161 pKa = 4.69LPIGDD166 pKa = 5.05FNTNQISYY174 pKa = 8.46TFATGGFAATLGLEE188 pKa = 4.38DD189 pKa = 5.96DD190 pKa = 4.38GTGDD194 pKa = 3.63IAPDD198 pKa = 3.34VHH200 pKa = 6.86GKK202 pKa = 10.3LSYY205 pKa = 8.91TGAWGGVFLSAVYY218 pKa = 10.68DD219 pKa = 3.59EE220 pKa = 4.89TFNAEE225 pKa = 4.16DD226 pKa = 3.5VTDD229 pKa = 4.91LFGIDD234 pKa = 5.57DD235 pKa = 4.92LDD237 pKa = 4.37LPAFFPFIPEE247 pKa = 3.84RR248 pKa = 11.84DD249 pKa = 3.3VDD251 pKa = 3.87GSNDD255 pKa = 3.23AFALKK260 pKa = 10.53AGLTLQDD267 pKa = 4.62LLVADD272 pKa = 4.2STLKK276 pKa = 10.71VDD278 pKa = 3.81GHH280 pKa = 6.08WASDD284 pKa = 3.44PTSYY288 pKa = 11.4AVISGFTSAGGILPSNATLEE308 pKa = 4.26NPTPGAFPLVLEE320 pKa = 4.45WSVGAGYY327 pKa = 8.38SQKK330 pKa = 10.28FNRR333 pKa = 11.84VTAGVSGRR341 pKa = 11.84YY342 pKa = 9.77GEE344 pKa = 4.26TFDD347 pKa = 3.88YY348 pKa = 11.27AFVAPEE354 pKa = 3.99VPGGFVFSSADD365 pKa = 3.19YY366 pKa = 8.63WAVVGDD372 pKa = 3.74IGYY375 pKa = 10.2DD376 pKa = 3.1LTANMNVLAEE386 pKa = 4.17VKK388 pKa = 10.08YY389 pKa = 10.88QNIDD393 pKa = 3.37FGGVASDD400 pKa = 4.04DD401 pKa = 3.68QTSGFLRR408 pKa = 11.84FQRR411 pKa = 11.84NFF413 pKa = 3.06
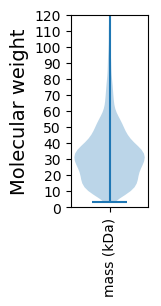
Molecular weight: 44.13 kDa
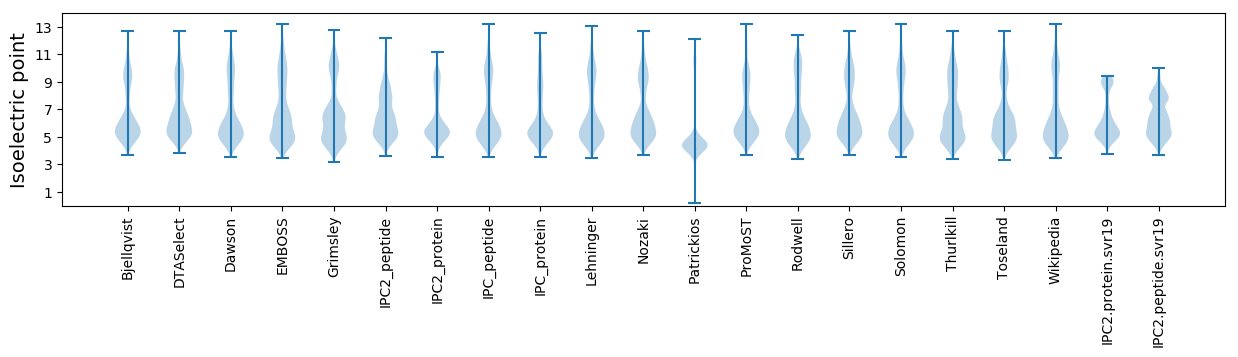
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B1Q233|A0A0B1Q233_9RHIZ DNA-3-methyladenine glycosidase OS=Aureimonas altamirensis OX=370622 GN=LA66_19850 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATTGGRR28 pKa = 11.84RR29 pKa = 11.84VIASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATTGGRR28 pKa = 11.84RR29 pKa = 11.84VIASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1115690 |
26 |
2158 |
317.0 |
34.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.011 ± 0.058 | 0.76 ± 0.012 |
5.846 ± 0.038 | 5.653 ± 0.04 |
3.785 ± 0.03 | 8.925 ± 0.033 |
1.973 ± 0.02 | 5.197 ± 0.028 |
2.525 ± 0.029 | 10.154 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.587 ± 0.021 | 2.335 ± 0.021 |
5.141 ± 0.027 | 2.973 ± 0.023 |
7.558 ± 0.036 | 5.344 ± 0.028 |
5.214 ± 0.029 | 7.604 ± 0.032 |
1.235 ± 0.013 | 2.178 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
