
Candidatus Izimaplasma sp. ZiA1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Tenericutes incertae sedis; Candidatus Izimaplasma; unclassified Candidatus Izimaplasma
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
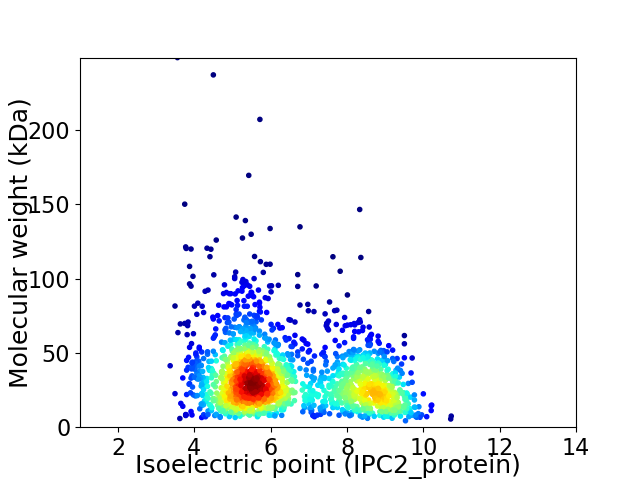
Virtual 2D-PAGE plot for 1828 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A1ZTL8|A0A2A1ZTL8_9BACT 50S ribosomal protein L11 OS=Candidatus Izimaplasma sp. ZiA1 OX=2024899 GN=rplK PE=3 SV=1
MM1 pKa = 7.8DD2 pKa = 6.7LIEE5 pKa = 4.49EE6 pKa = 4.9MIPEE10 pKa = 4.54ANVSTTYY17 pKa = 11.36DD18 pKa = 3.01MSSFQDD24 pKa = 3.89AIVEE28 pKa = 4.3MVAQTRR34 pKa = 11.84NGVLGVVAQEE44 pKa = 4.36LSGSTVIGGGTGSGVVYY61 pKa = 10.52KK62 pKa = 10.71KK63 pKa = 11.05VGDD66 pKa = 3.62VYY68 pKa = 11.45YY69 pKa = 10.7LVTNAHH75 pKa = 6.71VITEE79 pKa = 4.46DD80 pKa = 3.41VTDD83 pKa = 3.58TLGQVTGTEE92 pKa = 3.97LLDD95 pKa = 3.65NFVITYY101 pKa = 7.5EE102 pKa = 4.24KK103 pKa = 11.29NNLLFNINEE112 pKa = 4.29VTVVGYY118 pKa = 10.72DD119 pKa = 3.36LTTDD123 pKa = 4.28LAVLTFTSDD132 pKa = 2.92EE133 pKa = 4.21YY134 pKa = 11.36FSVIEE139 pKa = 4.69FGDD142 pKa = 3.7SYY144 pKa = 11.35AIEE147 pKa = 4.63PGQFVFAIGNPLGFNYY163 pKa = 10.58YY164 pKa = 8.62GTLTMGVISGTSRR177 pKa = 11.84YY178 pKa = 9.47LQDD181 pKa = 3.36GDD183 pKa = 4.34FDD185 pKa = 4.08ATLLQHH191 pKa = 7.1DD192 pKa = 4.4AAISPGNSGGALVDD206 pKa = 3.62INGKK210 pKa = 10.1LIGINNMKK218 pKa = 10.18IVDD221 pKa = 3.97ADD223 pKa = 3.54VTGIGFAIPSNTVQRR238 pKa = 11.84IVEE241 pKa = 4.2DD242 pKa = 4.23LEE244 pKa = 5.88DD245 pKa = 4.77DD246 pKa = 4.33GVVTRR251 pKa = 11.84PFLGIVTSAQVNQCGIDD268 pKa = 3.52YY269 pKa = 8.96GVCLLDD275 pKa = 5.27SEE277 pKa = 4.84SQPAIYY283 pKa = 10.01PNGAADD289 pKa = 3.71NAEE292 pKa = 4.24LQSGDD297 pKa = 3.75VIIGFKK303 pKa = 10.66KK304 pKa = 9.76IDD306 pKa = 3.32QEE308 pKa = 4.47EE309 pKa = 4.59YY310 pKa = 10.68FDD312 pKa = 3.95VFNFNDD318 pKa = 3.67LKK320 pKa = 11.08EE321 pKa = 5.18AILNCKK327 pKa = 10.21VGDD330 pKa = 3.91QIQIKK335 pKa = 8.71YY336 pKa = 10.12IRR338 pKa = 11.84DD339 pKa = 3.59NEE341 pKa = 4.22IKK343 pKa = 10.19EE344 pKa = 4.43SNIVTLGVHH353 pKa = 6.85PDD355 pKa = 3.49DD356 pKa = 4.49
MM1 pKa = 7.8DD2 pKa = 6.7LIEE5 pKa = 4.49EE6 pKa = 4.9MIPEE10 pKa = 4.54ANVSTTYY17 pKa = 11.36DD18 pKa = 3.01MSSFQDD24 pKa = 3.89AIVEE28 pKa = 4.3MVAQTRR34 pKa = 11.84NGVLGVVAQEE44 pKa = 4.36LSGSTVIGGGTGSGVVYY61 pKa = 10.52KK62 pKa = 10.71KK63 pKa = 11.05VGDD66 pKa = 3.62VYY68 pKa = 11.45YY69 pKa = 10.7LVTNAHH75 pKa = 6.71VITEE79 pKa = 4.46DD80 pKa = 3.41VTDD83 pKa = 3.58TLGQVTGTEE92 pKa = 3.97LLDD95 pKa = 3.65NFVITYY101 pKa = 7.5EE102 pKa = 4.24KK103 pKa = 11.29NNLLFNINEE112 pKa = 4.29VTVVGYY118 pKa = 10.72DD119 pKa = 3.36LTTDD123 pKa = 4.28LAVLTFTSDD132 pKa = 2.92EE133 pKa = 4.21YY134 pKa = 11.36FSVIEE139 pKa = 4.69FGDD142 pKa = 3.7SYY144 pKa = 11.35AIEE147 pKa = 4.63PGQFVFAIGNPLGFNYY163 pKa = 10.58YY164 pKa = 8.62GTLTMGVISGTSRR177 pKa = 11.84YY178 pKa = 9.47LQDD181 pKa = 3.36GDD183 pKa = 4.34FDD185 pKa = 4.08ATLLQHH191 pKa = 7.1DD192 pKa = 4.4AAISPGNSGGALVDD206 pKa = 3.62INGKK210 pKa = 10.1LIGINNMKK218 pKa = 10.18IVDD221 pKa = 3.97ADD223 pKa = 3.54VTGIGFAIPSNTVQRR238 pKa = 11.84IVEE241 pKa = 4.2DD242 pKa = 4.23LEE244 pKa = 5.88DD245 pKa = 4.77DD246 pKa = 4.33GVVTRR251 pKa = 11.84PFLGIVTSAQVNQCGIDD268 pKa = 3.52YY269 pKa = 8.96GVCLLDD275 pKa = 5.27SEE277 pKa = 4.84SQPAIYY283 pKa = 10.01PNGAADD289 pKa = 3.71NAEE292 pKa = 4.24LQSGDD297 pKa = 3.75VIIGFKK303 pKa = 10.66KK304 pKa = 9.76IDD306 pKa = 3.32QEE308 pKa = 4.47EE309 pKa = 4.59YY310 pKa = 10.68FDD312 pKa = 3.95VFNFNDD318 pKa = 3.67LKK320 pKa = 11.08EE321 pKa = 5.18AILNCKK327 pKa = 10.21VGDD330 pKa = 3.91QIQIKK335 pKa = 8.71YY336 pKa = 10.12IRR338 pKa = 11.84DD339 pKa = 3.59NEE341 pKa = 4.22IKK343 pKa = 10.19EE344 pKa = 4.43SNIVTLGVHH353 pKa = 6.85PDD355 pKa = 3.49DD356 pKa = 4.49
Molecular weight: 38.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A1ZNL1|A0A2A1ZNL1_9BACT Uncharacterized protein OS=Candidatus Izimaplasma sp. ZiA1 OX=2024899 GN=CI105_09305 PE=4 SV=1
MM1 pKa = 6.89PAAWKK6 pKa = 10.18GRR8 pKa = 11.84SGRR11 pKa = 11.84VWGKK15 pKa = 9.17RR16 pKa = 11.84GKK18 pKa = 9.4WPVKK22 pKa = 9.85PGLTFHH28 pKa = 7.31IDD30 pKa = 3.04DD31 pKa = 3.84SAYY34 pKa = 8.68FQYY37 pKa = 10.74RR38 pKa = 11.84RR39 pKa = 11.84IVRR42 pKa = 11.84SWRR45 pKa = 11.84IADD48 pKa = 3.48ARR50 pKa = 11.84SEE52 pKa = 4.25EE53 pKa = 4.2NPQNRR58 pKa = 11.84AWRR61 pKa = 11.84IRR63 pKa = 11.84SAKK66 pKa = 10.21KK67 pKa = 9.4MLKK70 pKa = 10.37CSDD73 pKa = 3.54RR74 pKa = 11.84ALSEE78 pKa = 4.01VRR80 pKa = 11.84GTNEE84 pKa = 4.04WISEE88 pKa = 4.08ANTFRR93 pKa = 11.84IIAYY97 pKa = 9.12LAAGGCEE104 pKa = 4.41VYY106 pKa = 10.66SAA108 pKa = 4.4
MM1 pKa = 6.89PAAWKK6 pKa = 10.18GRR8 pKa = 11.84SGRR11 pKa = 11.84VWGKK15 pKa = 9.17RR16 pKa = 11.84GKK18 pKa = 9.4WPVKK22 pKa = 9.85PGLTFHH28 pKa = 7.31IDD30 pKa = 3.04DD31 pKa = 3.84SAYY34 pKa = 8.68FQYY37 pKa = 10.74RR38 pKa = 11.84RR39 pKa = 11.84IVRR42 pKa = 11.84SWRR45 pKa = 11.84IADD48 pKa = 3.48ARR50 pKa = 11.84SEE52 pKa = 4.25EE53 pKa = 4.2NPQNRR58 pKa = 11.84AWRR61 pKa = 11.84IRR63 pKa = 11.84SAKK66 pKa = 10.21KK67 pKa = 9.4MLKK70 pKa = 10.37CSDD73 pKa = 3.54RR74 pKa = 11.84ALSEE78 pKa = 4.01VRR80 pKa = 11.84GTNEE84 pKa = 4.04WISEE88 pKa = 4.08ANTFRR93 pKa = 11.84IIAYY97 pKa = 9.12LAAGGCEE104 pKa = 4.41VYY106 pKa = 10.66SAA108 pKa = 4.4
Molecular weight: 12.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
575450 |
37 |
2306 |
314.8 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.072 ± 0.057 | 0.735 ± 0.018 |
5.99 ± 0.051 | 6.875 ± 0.053 |
4.957 ± 0.047 | 5.863 ± 0.055 |
1.625 ± 0.021 | 9.738 ± 0.069 |
8.397 ± 0.071 | 9.837 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.028 | 6.334 ± 0.07 |
2.619 ± 0.03 | 2.367 ± 0.026 |
3.183 ± 0.038 | 6.427 ± 0.05 |
5.497 ± 0.043 | 6.866 ± 0.047 |
0.543 ± 0.014 | 4.588 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
