
Thalassotalea sp. HSM 43
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Colwelliaceae; Thalassotalea; unclassified Thalassotalea
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
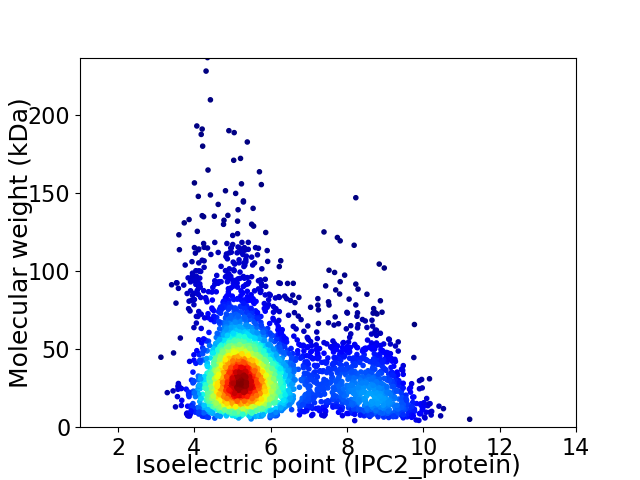
Virtual 2D-PAGE plot for 3411 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P7JJL3|A0A4P7JJL3_9GAMM Uncharacterized protein OS=Thalassotalea sp. HSM 43 OX=2552945 GN=E2K93_01510 PE=4 SV=1
MM1 pKa = 7.57SKK3 pKa = 10.44NLNKK7 pKa = 10.33SYY9 pKa = 10.79LALLVASALSLTACGGGGGGGGTEE33 pKa = 4.23KK34 pKa = 10.99KK35 pKa = 10.42EE36 pKa = 4.13IEE38 pKa = 4.56PPTNNAPTISGSPSLSINEE57 pKa = 3.97NQSYY61 pKa = 10.78SFSANGADD69 pKa = 3.82SDD71 pKa = 4.39GDD73 pKa = 3.75ALTYY77 pKa = 10.61SIANMPQWAAFDD89 pKa = 4.15SATGTLTGTPGNADD103 pKa = 2.93VGTFTGITISVSDD116 pKa = 3.95GTDD119 pKa = 3.08SSSLSPFDD127 pKa = 5.22IIVNDD132 pKa = 3.75IFALQGSATLEE143 pKa = 4.16VTVGEE148 pKa = 4.62TNVFTPTLVDD158 pKa = 3.83HH159 pKa = 6.87VDD161 pKa = 3.12ADD163 pKa = 4.14FSYY166 pKa = 10.92SIDD169 pKa = 3.68GAPSFAGFSFDD180 pKa = 3.62AATGQVSVTPAADD193 pKa = 4.64DD194 pKa = 3.38IGSFNITITASNGVNSASHH213 pKa = 6.33NMTLTISPGSISVAGKK229 pKa = 10.7VIDD232 pKa = 4.54GYY234 pKa = 10.91ISGAHH239 pKa = 5.75VFLDD243 pKa = 3.64VNNNMMFDD251 pKa = 4.05DD252 pKa = 4.72NEE254 pKa = 4.21PNALSNNAGEE264 pKa = 4.44FTLDD268 pKa = 3.61VPGTLIDD275 pKa = 4.79SIPTSTLVAEE285 pKa = 4.98LGSGQGVDD293 pKa = 3.81ADD295 pKa = 3.92DD296 pKa = 3.87MSRR299 pKa = 11.84FDD301 pKa = 5.27DD302 pKa = 4.78DD303 pKa = 3.73FAARR307 pKa = 11.84PIKK310 pKa = 9.93MMSYY314 pKa = 10.17PIRR317 pKa = 11.84DD318 pKa = 3.94LTNMSNGLTAYY329 pKa = 7.37ITPFTHH335 pKa = 7.74LIHH338 pKa = 7.05EE339 pKa = 5.29EE340 pKa = 3.86ILSAAGGDD348 pKa = 3.54LSLITDD354 pKa = 3.46QEE356 pKa = 4.67LYY358 pKa = 11.37NLIKK362 pKa = 10.74DD363 pKa = 3.98SAVDD367 pKa = 4.77RR368 pKa = 11.84INGLYY373 pKa = 10.59SVDD376 pKa = 4.66LDD378 pKa = 4.25LATSDD383 pKa = 3.91FFAAEE388 pKa = 3.97VPINSRR394 pKa = 11.84NDD396 pKa = 3.0IASVAKK402 pKa = 10.48AVLNDD407 pKa = 4.09LQDD410 pKa = 3.41QVNDD414 pKa = 3.54SDD416 pKa = 4.06QDD418 pKa = 3.76GFVNAEE424 pKa = 4.2DD425 pKa = 4.55DD426 pKa = 4.14LPFNSEE432 pKa = 4.41YY433 pKa = 10.48ISDD436 pKa = 3.76IDD438 pKa = 3.82QDD440 pKa = 4.03GLGDD444 pKa = 4.17CVRR447 pKa = 11.84TIPGKK452 pKa = 10.4SCVEE456 pKa = 3.99EE457 pKa = 6.17DD458 pKa = 5.23DD459 pKa = 6.62DD460 pKa = 5.98IDD462 pKa = 5.19GDD464 pKa = 4.29NITNADD470 pKa = 4.0DD471 pKa = 4.35PEE473 pKa = 4.36PTVPAGLAVFKK484 pKa = 9.66PVPEE488 pKa = 4.18MNVPGYY494 pKa = 8.65EE495 pKa = 3.96MCVGLVNQDD504 pKa = 3.71KK505 pKa = 10.81VLCLDD510 pKa = 3.97NYY512 pKa = 9.74QWSILDD518 pKa = 4.01SIKK521 pKa = 11.07GEE523 pKa = 4.12PLPIDD528 pKa = 3.43TNSIVKK534 pKa = 10.52SGDD537 pKa = 2.96SAAAVGTCAVANNEE551 pKa = 4.23VYY553 pKa = 10.84CLDD556 pKa = 3.07GKK558 pKa = 10.07MRR560 pKa = 11.84FDD562 pKa = 3.93FAGEE566 pKa = 4.14NPVSLNPIALEE577 pKa = 3.74MGYY580 pKa = 11.17LSMCYY585 pKa = 9.95LDD587 pKa = 3.65QTGVKK592 pKa = 9.64CFGGIEE598 pKa = 3.99EE599 pKa = 4.92SMPEE603 pKa = 4.08SLVNASALALTSGNACAISEE623 pKa = 4.36VTDD626 pKa = 3.47NEE628 pKa = 4.32QTSTAVSCWGSDD640 pKa = 3.24SYY642 pKa = 12.09GLTTGVPVLSNPTQLFSNMTTICALDD668 pKa = 3.58DD669 pKa = 3.66TGIKK673 pKa = 10.1CWGNPDD679 pKa = 3.56YY680 pKa = 11.49NLDD683 pKa = 4.01KK684 pKa = 10.92VDD686 pKa = 3.69HH687 pKa = 6.9ASTKK691 pKa = 10.37LSNIHH696 pKa = 5.89LTGDD700 pKa = 3.59SLCFIDD706 pKa = 5.23EE707 pKa = 4.22QTPVCYY713 pKa = 10.47GNSDD717 pKa = 3.57QPTPISADD725 pKa = 2.96KK726 pKa = 10.89DD727 pKa = 3.84YY728 pKa = 11.45FDD730 pKa = 5.64ADD732 pKa = 3.56SGEE735 pKa = 4.78LIVAHH740 pKa = 6.52GEE742 pKa = 4.13FIAPFEE748 pKa = 4.23EE749 pKa = 4.37QLTKK753 pKa = 10.79YY754 pKa = 10.66VEE756 pKa = 4.63GVHH759 pKa = 4.77QQRR762 pKa = 11.84VLLPEE767 pKa = 3.75VDD769 pKa = 4.53FEE771 pKa = 4.4QGQTATEE778 pKa = 4.17VFRR781 pKa = 11.84LRR783 pKa = 11.84NYY785 pKa = 7.95TKK787 pKa = 9.15NTCLISDD794 pKa = 4.54GEE796 pKa = 4.29ISCDD800 pKa = 3.26SQRR803 pKa = 11.84YY804 pKa = 8.73CGLDD808 pKa = 3.13NGEE811 pKa = 4.26WQCQTVEE818 pKa = 4.48FNFDD822 pKa = 4.21DD823 pKa = 3.65IDD825 pKa = 4.54FEE827 pKa = 5.24VKK829 pKa = 9.02TQINNSNTVCAIGALKK845 pKa = 9.95AHH847 pKa = 6.47CVGAGFNGEE856 pKa = 3.86IKK858 pKa = 10.94LNN860 pKa = 3.7
MM1 pKa = 7.57SKK3 pKa = 10.44NLNKK7 pKa = 10.33SYY9 pKa = 10.79LALLVASALSLTACGGGGGGGGTEE33 pKa = 4.23KK34 pKa = 10.99KK35 pKa = 10.42EE36 pKa = 4.13IEE38 pKa = 4.56PPTNNAPTISGSPSLSINEE57 pKa = 3.97NQSYY61 pKa = 10.78SFSANGADD69 pKa = 3.82SDD71 pKa = 4.39GDD73 pKa = 3.75ALTYY77 pKa = 10.61SIANMPQWAAFDD89 pKa = 4.15SATGTLTGTPGNADD103 pKa = 2.93VGTFTGITISVSDD116 pKa = 3.95GTDD119 pKa = 3.08SSSLSPFDD127 pKa = 5.22IIVNDD132 pKa = 3.75IFALQGSATLEE143 pKa = 4.16VTVGEE148 pKa = 4.62TNVFTPTLVDD158 pKa = 3.83HH159 pKa = 6.87VDD161 pKa = 3.12ADD163 pKa = 4.14FSYY166 pKa = 10.92SIDD169 pKa = 3.68GAPSFAGFSFDD180 pKa = 3.62AATGQVSVTPAADD193 pKa = 4.64DD194 pKa = 3.38IGSFNITITASNGVNSASHH213 pKa = 6.33NMTLTISPGSISVAGKK229 pKa = 10.7VIDD232 pKa = 4.54GYY234 pKa = 10.91ISGAHH239 pKa = 5.75VFLDD243 pKa = 3.64VNNNMMFDD251 pKa = 4.05DD252 pKa = 4.72NEE254 pKa = 4.21PNALSNNAGEE264 pKa = 4.44FTLDD268 pKa = 3.61VPGTLIDD275 pKa = 4.79SIPTSTLVAEE285 pKa = 4.98LGSGQGVDD293 pKa = 3.81ADD295 pKa = 3.92DD296 pKa = 3.87MSRR299 pKa = 11.84FDD301 pKa = 5.27DD302 pKa = 4.78DD303 pKa = 3.73FAARR307 pKa = 11.84PIKK310 pKa = 9.93MMSYY314 pKa = 10.17PIRR317 pKa = 11.84DD318 pKa = 3.94LTNMSNGLTAYY329 pKa = 7.37ITPFTHH335 pKa = 7.74LIHH338 pKa = 7.05EE339 pKa = 5.29EE340 pKa = 3.86ILSAAGGDD348 pKa = 3.54LSLITDD354 pKa = 3.46QEE356 pKa = 4.67LYY358 pKa = 11.37NLIKK362 pKa = 10.74DD363 pKa = 3.98SAVDD367 pKa = 4.77RR368 pKa = 11.84INGLYY373 pKa = 10.59SVDD376 pKa = 4.66LDD378 pKa = 4.25LATSDD383 pKa = 3.91FFAAEE388 pKa = 3.97VPINSRR394 pKa = 11.84NDD396 pKa = 3.0IASVAKK402 pKa = 10.48AVLNDD407 pKa = 4.09LQDD410 pKa = 3.41QVNDD414 pKa = 3.54SDD416 pKa = 4.06QDD418 pKa = 3.76GFVNAEE424 pKa = 4.2DD425 pKa = 4.55DD426 pKa = 4.14LPFNSEE432 pKa = 4.41YY433 pKa = 10.48ISDD436 pKa = 3.76IDD438 pKa = 3.82QDD440 pKa = 4.03GLGDD444 pKa = 4.17CVRR447 pKa = 11.84TIPGKK452 pKa = 10.4SCVEE456 pKa = 3.99EE457 pKa = 6.17DD458 pKa = 5.23DD459 pKa = 6.62DD460 pKa = 5.98IDD462 pKa = 5.19GDD464 pKa = 4.29NITNADD470 pKa = 4.0DD471 pKa = 4.35PEE473 pKa = 4.36PTVPAGLAVFKK484 pKa = 9.66PVPEE488 pKa = 4.18MNVPGYY494 pKa = 8.65EE495 pKa = 3.96MCVGLVNQDD504 pKa = 3.71KK505 pKa = 10.81VLCLDD510 pKa = 3.97NYY512 pKa = 9.74QWSILDD518 pKa = 4.01SIKK521 pKa = 11.07GEE523 pKa = 4.12PLPIDD528 pKa = 3.43TNSIVKK534 pKa = 10.52SGDD537 pKa = 2.96SAAAVGTCAVANNEE551 pKa = 4.23VYY553 pKa = 10.84CLDD556 pKa = 3.07GKK558 pKa = 10.07MRR560 pKa = 11.84FDD562 pKa = 3.93FAGEE566 pKa = 4.14NPVSLNPIALEE577 pKa = 3.74MGYY580 pKa = 11.17LSMCYY585 pKa = 9.95LDD587 pKa = 3.65QTGVKK592 pKa = 9.64CFGGIEE598 pKa = 3.99EE599 pKa = 4.92SMPEE603 pKa = 4.08SLVNASALALTSGNACAISEE623 pKa = 4.36VTDD626 pKa = 3.47NEE628 pKa = 4.32QTSTAVSCWGSDD640 pKa = 3.24SYY642 pKa = 12.09GLTTGVPVLSNPTQLFSNMTTICALDD668 pKa = 3.58DD669 pKa = 3.66TGIKK673 pKa = 10.1CWGNPDD679 pKa = 3.56YY680 pKa = 11.49NLDD683 pKa = 4.01KK684 pKa = 10.92VDD686 pKa = 3.69HH687 pKa = 6.9ASTKK691 pKa = 10.37LSNIHH696 pKa = 5.89LTGDD700 pKa = 3.59SLCFIDD706 pKa = 5.23EE707 pKa = 4.22QTPVCYY713 pKa = 10.47GNSDD717 pKa = 3.57QPTPISADD725 pKa = 2.96KK726 pKa = 10.89DD727 pKa = 3.84YY728 pKa = 11.45FDD730 pKa = 5.64ADD732 pKa = 3.56SGEE735 pKa = 4.78LIVAHH740 pKa = 6.52GEE742 pKa = 4.13FIAPFEE748 pKa = 4.23EE749 pKa = 4.37QLTKK753 pKa = 10.79YY754 pKa = 10.66VEE756 pKa = 4.63GVHH759 pKa = 4.77QQRR762 pKa = 11.84VLLPEE767 pKa = 3.75VDD769 pKa = 4.53FEE771 pKa = 4.4QGQTATEE778 pKa = 4.17VFRR781 pKa = 11.84LRR783 pKa = 11.84NYY785 pKa = 7.95TKK787 pKa = 9.15NTCLISDD794 pKa = 4.54GEE796 pKa = 4.29ISCDD800 pKa = 3.26SQRR803 pKa = 11.84YY804 pKa = 8.73CGLDD808 pKa = 3.13NGEE811 pKa = 4.26WQCQTVEE818 pKa = 4.48FNFDD822 pKa = 4.21DD823 pKa = 3.65IDD825 pKa = 4.54FEE827 pKa = 5.24VKK829 pKa = 9.02TQINNSNTVCAIGALKK845 pKa = 9.95AHH847 pKa = 6.47CVGAGFNGEE856 pKa = 3.86IKK858 pKa = 10.94LNN860 pKa = 3.7
Molecular weight: 91.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P7JQM4|A0A4P7JQM4_9GAMM UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2 6-diaminopimelate ligase OS=Thalassotalea sp. HSM 43 OX=2552945 GN=murE PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84AKK41 pKa = 10.57LSSS44 pKa = 3.31
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84AKK41 pKa = 10.57LSSS44 pKa = 3.31
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1170031 |
38 |
2158 |
343.0 |
38.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.691 ± 0.044 | 0.995 ± 0.014 |
6.223 ± 0.04 | 5.791 ± 0.036 |
4.292 ± 0.025 | 6.731 ± 0.042 |
2.263 ± 0.021 | 6.527 ± 0.03 |
5.386 ± 0.038 | 9.743 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.642 ± 0.019 | 4.692 ± 0.029 |
3.705 ± 0.021 | 5.077 ± 0.041 |
4.117 ± 0.033 | 6.54 ± 0.032 |
5.162 ± 0.033 | 6.917 ± 0.034 |
1.22 ± 0.016 | 3.287 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
