
Butyrivibrio virus Idris
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
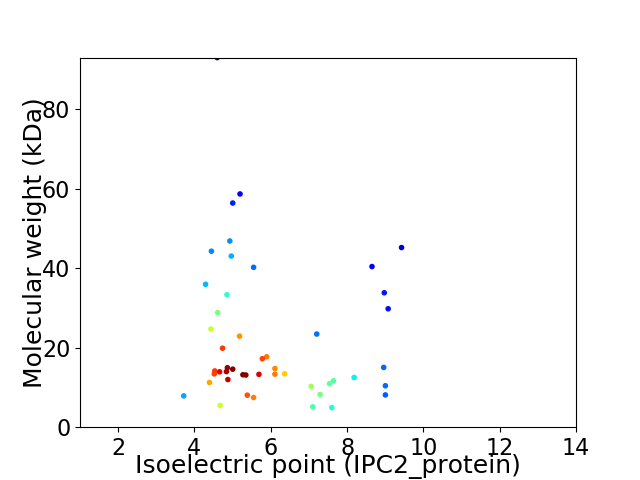
Virtual 2D-PAGE plot for 48 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6H1MY30|A0A6H1MY30_9CAUD Uncharacterized protein OS=Butyrivibrio virus Idris OX=2696360 PE=4 SV=1
MM1 pKa = 7.5PVWKK5 pKa = 9.88YY6 pKa = 11.4DD7 pKa = 3.24EE8 pKa = 4.66AAISRR13 pKa = 11.84AEE15 pKa = 3.98YY16 pKa = 9.96EE17 pKa = 4.25KK18 pKa = 11.54NEE20 pKa = 4.45AIYY23 pKa = 10.89SEE25 pKa = 4.51LVVMIEE31 pKa = 3.75QQAANDD37 pKa = 3.86SVYY40 pKa = 11.27GDD42 pKa = 4.06VLLQQADD49 pKa = 3.68AQATLAEE56 pKa = 4.15QDD58 pKa = 3.59EE59 pKa = 4.63VLADD63 pKa = 3.45ILLTVQGVV71 pKa = 3.15
MM1 pKa = 7.5PVWKK5 pKa = 9.88YY6 pKa = 11.4DD7 pKa = 3.24EE8 pKa = 4.66AAISRR13 pKa = 11.84AEE15 pKa = 3.98YY16 pKa = 9.96EE17 pKa = 4.25KK18 pKa = 11.54NEE20 pKa = 4.45AIYY23 pKa = 10.89SEE25 pKa = 4.51LVVMIEE31 pKa = 3.75QQAANDD37 pKa = 3.86SVYY40 pKa = 11.27GDD42 pKa = 4.06VLLQQADD49 pKa = 3.68AQATLAEE56 pKa = 4.15QDD58 pKa = 3.59EE59 pKa = 4.63VLADD63 pKa = 3.45ILLTVQGVV71 pKa = 3.15
Molecular weight: 7.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H1MY30|A0A6H1MY30_9CAUD Uncharacterized protein OS=Butyrivibrio virus Idris OX=2696360 PE=4 SV=1
MM1 pKa = 7.21KK2 pKa = 9.67TYY4 pKa = 10.8KK5 pKa = 10.35HH6 pKa = 6.32LLEE9 pKa = 4.87QIADD13 pKa = 3.53KK14 pKa = 11.5DD15 pKa = 3.98NVRR18 pKa = 11.84KK19 pKa = 10.12AILNASKK26 pKa = 10.56RR27 pKa = 11.84KK28 pKa = 8.87RR29 pKa = 11.84HH30 pKa = 5.85RR31 pKa = 11.84KK32 pKa = 8.88DD33 pKa = 3.02VKK35 pKa = 10.61QVIEE39 pKa = 4.49NIDD42 pKa = 3.31YY43 pKa = 10.63HH44 pKa = 7.75VEE46 pKa = 4.18VVHH49 pKa = 6.78NMLLAGSYY57 pKa = 9.03KK58 pKa = 10.45AHH60 pKa = 6.65VDD62 pKa = 3.37APCIVNEE69 pKa = 4.33GTHH72 pKa = 5.63HH73 pKa = 6.33KK74 pKa = 10.07VRR76 pKa = 11.84RR77 pKa = 11.84IRR79 pKa = 11.84KK80 pKa = 7.5PHH82 pKa = 5.59YY83 pKa = 10.02KK84 pKa = 9.19YY85 pKa = 10.74DD86 pKa = 3.9QIIHH90 pKa = 6.03HH91 pKa = 6.92CIIQVLQPILTAPMYY106 pKa = 9.71EE107 pKa = 4.24YY108 pKa = 11.23SCGSIPKK115 pKa = 10.12RR116 pKa = 11.84GAHH119 pKa = 4.87YY120 pKa = 10.2GKK122 pKa = 10.16RR123 pKa = 11.84RR124 pKa = 11.84LEE126 pKa = 3.42KK127 pKa = 10.05WIRR130 pKa = 11.84NDD132 pKa = 3.33AKK134 pKa = 9.11GTKK137 pKa = 9.62YY138 pKa = 10.16VFKK141 pKa = 10.39MDD143 pKa = 2.78IKK145 pKa = 10.73HH146 pKa = 6.12FYY148 pKa = 10.53EE149 pKa = 5.45SVDD152 pKa = 3.49QDD154 pKa = 3.78VLKK157 pKa = 11.38SMLKK161 pKa = 10.64AKK163 pKa = 10.32IKK165 pKa = 10.16DD166 pKa = 3.18WRR168 pKa = 11.84ALEE171 pKa = 4.68LIYY174 pKa = 10.25TVIDD178 pKa = 3.56SCEE181 pKa = 3.85KK182 pKa = 10.58GLPLGNYY189 pKa = 6.78TSQWFANFMLTPLDD203 pKa = 4.06HH204 pKa = 6.69YY205 pKa = 11.05IKK207 pKa = 10.22EE208 pKa = 4.13QLHH211 pKa = 5.78AKK213 pKa = 10.29YY214 pKa = 10.8YY215 pKa = 8.19MRR217 pKa = 11.84YY218 pKa = 8.23MDD220 pKa = 6.36DD221 pKa = 3.37IVILGGNKK229 pKa = 9.47KK230 pKa = 9.03EE231 pKa = 3.61LHH233 pKa = 6.72KK234 pKa = 10.34IHH236 pKa = 6.88KK237 pKa = 10.24AIEE240 pKa = 4.45KK241 pKa = 9.06YY242 pKa = 10.54LSEE245 pKa = 4.22RR246 pKa = 11.84LHH248 pKa = 7.91LRR250 pKa = 11.84IKK252 pKa = 10.32EE253 pKa = 3.76NWQVFRR259 pKa = 11.84LVYY262 pKa = 9.54TDD264 pKa = 3.08RR265 pKa = 11.84NGRR268 pKa = 11.84QRR270 pKa = 11.84GRR272 pKa = 11.84SLDD275 pKa = 3.5FMGWQFYY282 pKa = 10.25RR283 pKa = 11.84EE284 pKa = 3.94KK285 pKa = 10.33TILRR289 pKa = 11.84EE290 pKa = 4.11SIYY293 pKa = 10.91LRR295 pKa = 11.84IVRR298 pKa = 11.84KK299 pKa = 9.15ARR301 pKa = 11.84HH302 pKa = 5.31VGKK305 pKa = 8.76HH306 pKa = 4.3TTIQGAQGMISYY318 pKa = 8.93MGYY321 pKa = 9.7IKK323 pKa = 9.8HH324 pKa = 6.04TDD326 pKa = 2.91CYY328 pKa = 9.57GTYY331 pKa = 9.5RR332 pKa = 11.84DD333 pKa = 3.92YY334 pKa = 11.15IRR336 pKa = 11.84PYY338 pKa = 10.86VNIGKK343 pKa = 9.6LKK345 pKa = 10.57KK346 pKa = 9.2FTSKK350 pKa = 10.33RR351 pKa = 11.84AKK353 pKa = 10.42KK354 pKa = 9.95GVLKK358 pKa = 11.1NGVEE362 pKa = 4.04KK363 pKa = 10.65SDD365 pKa = 3.62RR366 pKa = 11.84NADD369 pKa = 3.4NTASSTRR376 pKa = 11.84PRR378 pKa = 11.84QQPP381 pKa = 2.92
MM1 pKa = 7.21KK2 pKa = 9.67TYY4 pKa = 10.8KK5 pKa = 10.35HH6 pKa = 6.32LLEE9 pKa = 4.87QIADD13 pKa = 3.53KK14 pKa = 11.5DD15 pKa = 3.98NVRR18 pKa = 11.84KK19 pKa = 10.12AILNASKK26 pKa = 10.56RR27 pKa = 11.84KK28 pKa = 8.87RR29 pKa = 11.84HH30 pKa = 5.85RR31 pKa = 11.84KK32 pKa = 8.88DD33 pKa = 3.02VKK35 pKa = 10.61QVIEE39 pKa = 4.49NIDD42 pKa = 3.31YY43 pKa = 10.63HH44 pKa = 7.75VEE46 pKa = 4.18VVHH49 pKa = 6.78NMLLAGSYY57 pKa = 9.03KK58 pKa = 10.45AHH60 pKa = 6.65VDD62 pKa = 3.37APCIVNEE69 pKa = 4.33GTHH72 pKa = 5.63HH73 pKa = 6.33KK74 pKa = 10.07VRR76 pKa = 11.84RR77 pKa = 11.84IRR79 pKa = 11.84KK80 pKa = 7.5PHH82 pKa = 5.59YY83 pKa = 10.02KK84 pKa = 9.19YY85 pKa = 10.74DD86 pKa = 3.9QIIHH90 pKa = 6.03HH91 pKa = 6.92CIIQVLQPILTAPMYY106 pKa = 9.71EE107 pKa = 4.24YY108 pKa = 11.23SCGSIPKK115 pKa = 10.12RR116 pKa = 11.84GAHH119 pKa = 4.87YY120 pKa = 10.2GKK122 pKa = 10.16RR123 pKa = 11.84RR124 pKa = 11.84LEE126 pKa = 3.42KK127 pKa = 10.05WIRR130 pKa = 11.84NDD132 pKa = 3.33AKK134 pKa = 9.11GTKK137 pKa = 9.62YY138 pKa = 10.16VFKK141 pKa = 10.39MDD143 pKa = 2.78IKK145 pKa = 10.73HH146 pKa = 6.12FYY148 pKa = 10.53EE149 pKa = 5.45SVDD152 pKa = 3.49QDD154 pKa = 3.78VLKK157 pKa = 11.38SMLKK161 pKa = 10.64AKK163 pKa = 10.32IKK165 pKa = 10.16DD166 pKa = 3.18WRR168 pKa = 11.84ALEE171 pKa = 4.68LIYY174 pKa = 10.25TVIDD178 pKa = 3.56SCEE181 pKa = 3.85KK182 pKa = 10.58GLPLGNYY189 pKa = 6.78TSQWFANFMLTPLDD203 pKa = 4.06HH204 pKa = 6.69YY205 pKa = 11.05IKK207 pKa = 10.22EE208 pKa = 4.13QLHH211 pKa = 5.78AKK213 pKa = 10.29YY214 pKa = 10.8YY215 pKa = 8.19MRR217 pKa = 11.84YY218 pKa = 8.23MDD220 pKa = 6.36DD221 pKa = 3.37IVILGGNKK229 pKa = 9.47KK230 pKa = 9.03EE231 pKa = 3.61LHH233 pKa = 6.72KK234 pKa = 10.34IHH236 pKa = 6.88KK237 pKa = 10.24AIEE240 pKa = 4.45KK241 pKa = 9.06YY242 pKa = 10.54LSEE245 pKa = 4.22RR246 pKa = 11.84LHH248 pKa = 7.91LRR250 pKa = 11.84IKK252 pKa = 10.32EE253 pKa = 3.76NWQVFRR259 pKa = 11.84LVYY262 pKa = 9.54TDD264 pKa = 3.08RR265 pKa = 11.84NGRR268 pKa = 11.84QRR270 pKa = 11.84GRR272 pKa = 11.84SLDD275 pKa = 3.5FMGWQFYY282 pKa = 10.25RR283 pKa = 11.84EE284 pKa = 3.94KK285 pKa = 10.33TILRR289 pKa = 11.84EE290 pKa = 4.11SIYY293 pKa = 10.91LRR295 pKa = 11.84IVRR298 pKa = 11.84KK299 pKa = 9.15ARR301 pKa = 11.84HH302 pKa = 5.31VGKK305 pKa = 8.76HH306 pKa = 4.3TTIQGAQGMISYY318 pKa = 8.93MGYY321 pKa = 9.7IKK323 pKa = 9.8HH324 pKa = 6.04TDD326 pKa = 2.91CYY328 pKa = 9.57GTYY331 pKa = 9.5RR332 pKa = 11.84DD333 pKa = 3.92YY334 pKa = 11.15IRR336 pKa = 11.84PYY338 pKa = 10.86VNIGKK343 pKa = 9.6LKK345 pKa = 10.57KK346 pKa = 9.2FTSKK350 pKa = 10.33RR351 pKa = 11.84AKK353 pKa = 10.42KK354 pKa = 9.95GVLKK358 pKa = 11.1NGVEE362 pKa = 4.04KK363 pKa = 10.65SDD365 pKa = 3.62RR366 pKa = 11.84NADD369 pKa = 3.4NTASSTRR376 pKa = 11.84PRR378 pKa = 11.84QQPP381 pKa = 2.92
Molecular weight: 45.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9576 |
47 |
882 |
199.5 |
22.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.91 ± 0.697 | 1.138 ± 0.147 |
5.994 ± 0.233 | 7.498 ± 0.466 |
2.955 ± 0.22 | 6.381 ± 0.346 |
1.431 ± 0.239 | 6.422 ± 0.35 |
7.268 ± 0.434 | 8.229 ± 0.361 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.318 ± 0.117 | 4.971 ± 0.282 |
2.642 ± 0.179 | 4.522 ± 0.211 |
4.313 ± 0.417 | 4.845 ± 0.252 |
6.704 ± 0.384 | 6.683 ± 0.326 |
1.117 ± 0.146 | 4.657 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
