
Dokdonella koreensis DS-123
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Rhodanobacteraceae; Dokdonella; Dokdonella koreensis
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
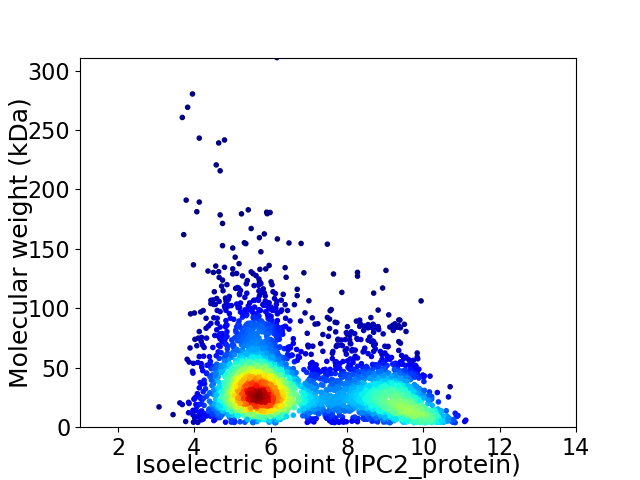
Virtual 2D-PAGE plot for 3774 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160DXW4|A0A160DXW4_9GAMM Uncharacterized protein OS=Dokdonella koreensis DS-123 OX=1300342 GN=I596_3599 PE=4 SV=1
MM1 pKa = 7.39TMQSRR6 pKa = 11.84AARR9 pKa = 11.84AAVSLCVMVVALAAGALGAQPATGPLQFSAPVSAPFVEE47 pKa = 4.87TKK49 pKa = 10.4PVVANPFSEE58 pKa = 4.34AARR61 pKa = 11.84PVSPRR66 pKa = 11.84AFSTEE71 pKa = 3.73FPAEE75 pKa = 4.03VEE77 pKa = 4.39PNGTPATATPLGAATYY93 pKa = 11.27VRR95 pKa = 11.84MQGNLFPAADD105 pKa = 3.25VDD107 pKa = 4.31YY108 pKa = 11.09FSFAAQAGDD117 pKa = 4.06RR118 pKa = 11.84IYY120 pKa = 11.3AAVITAANTSTSADD134 pKa = 3.53SQLNLLDD141 pKa = 3.82TDD143 pKa = 3.78GTTVLEE149 pKa = 4.51FDD151 pKa = 5.18DD152 pKa = 6.03DD153 pKa = 4.66DD154 pKa = 5.92GSFAATSSSIAGTVIPADD172 pKa = 3.28GTYY175 pKa = 10.21YY176 pKa = 10.97LSVNHH181 pKa = 5.78FTAAQTLRR189 pKa = 11.84PYY191 pKa = 9.28EE192 pKa = 3.78LHH194 pKa = 6.69VRR196 pKa = 11.84IQRR199 pKa = 11.84GSPTPEE205 pKa = 3.74VEE207 pKa = 4.98SNDD210 pKa = 3.75TPGTANPLPANGWVSGTRR228 pKa = 11.84NPPAATEE235 pKa = 3.98QDD237 pKa = 3.29WYY239 pKa = 11.18AFTANAGDD247 pKa = 4.02TVFLSLDD254 pKa = 3.77ADD256 pKa = 3.89PEE258 pKa = 4.32RR259 pKa = 11.84DD260 pKa = 3.4GVQWDD265 pKa = 3.61GRR267 pKa = 11.84LGIALFGDD275 pKa = 4.35AANQILVVNDD285 pKa = 3.69TSSGAGNPLSEE296 pKa = 4.46ALVFTIKK303 pKa = 9.79TAGTYY308 pKa = 9.75FAFVDD313 pKa = 3.85SATAAVGGPTATYY326 pKa = 8.87HH327 pKa = 6.62LSVSVQPPPTPMPLAACTTYY347 pKa = 10.85TSTDD351 pKa = 3.19VPKK354 pKa = 10.5TIGPANGLVSSLITVPGNPRR374 pKa = 11.84IGDD377 pKa = 3.93LNVEE381 pKa = 3.99IQLNHH386 pKa = 6.73ALMADD391 pKa = 3.52VDD393 pKa = 3.68AHH395 pKa = 6.32LRR397 pKa = 11.84SPSGNDD403 pKa = 2.46IGLFTDD409 pKa = 3.65IGAAATGGQAQMDD422 pKa = 4.0VVFDD426 pKa = 4.49DD427 pKa = 4.58EE428 pKa = 4.87AAIPPTFTALRR439 pKa = 11.84NMALKK444 pKa = 10.66PEE446 pKa = 4.26LNYY449 pKa = 10.57RR450 pKa = 11.84LSWFDD455 pKa = 3.41GEE457 pKa = 4.52NAGGTWTLDD466 pKa = 3.94LRR468 pKa = 11.84DD469 pKa = 4.39DD470 pKa = 4.06VNNASGGTLTSWSLRR485 pKa = 11.84ICEE488 pKa = 4.53PPPPPTCGPGFVQTTVYY505 pKa = 9.77STDD508 pKa = 3.92FEE510 pKa = 4.57SGAAGFTHH518 pKa = 7.23SGTQDD523 pKa = 2.74EE524 pKa = 4.98WEE526 pKa = 4.66LGLPATVATTTANPVAAFNSCASGVNCWKK555 pKa = 9.61TDD557 pKa = 3.3LDD559 pKa = 3.48NTYY562 pKa = 10.8NISSNQNLTSPSINLGGFSPPIVVRR587 pKa = 11.84WSHH590 pKa = 5.1QYY592 pKa = 10.56QMEE595 pKa = 4.34SASFDD600 pKa = 3.92LYY602 pKa = 11.36SVDD605 pKa = 3.36LRR607 pKa = 11.84EE608 pKa = 4.21VGGANPVRR616 pKa = 11.84LFEE619 pKa = 4.35WLDD622 pKa = 3.3ATMTDD627 pKa = 3.2AVGNPTVNIPARR639 pKa = 11.84AGWSQVVTRR648 pKa = 11.84ADD650 pKa = 3.17ALAGQNVEE658 pKa = 4.9LNFNLTSDD666 pKa = 3.38NTVNFGGVAVDD677 pKa = 4.07DD678 pKa = 4.53VSVTACRR685 pKa = 11.84VAIADD690 pKa = 4.0LSITKK695 pKa = 9.61TDD697 pKa = 3.45GVTTAVPGGSVTYY710 pKa = 10.02TITASNAGPDD720 pKa = 3.77ADD722 pKa = 4.15TAATVADD729 pKa = 4.52TFPAALTCTWTCVGAGGGTCTAAGSGNIADD759 pKa = 4.33TVNLPSGGSVTYY771 pKa = 8.84TASCAIGPSVTGTLSNTATVTASVADD797 pKa = 4.17PNPANNSATDD807 pKa = 3.63TDD809 pKa = 4.28TLTPQADD816 pKa = 3.73LAITKK821 pKa = 9.28TDD823 pKa = 3.06GVTSVIQGGSTTYY836 pKa = 10.62TITASNAGPSNASGATVTDD855 pKa = 4.01TFPASLTCTWTCVGAGGGTCTAAGSGNIADD885 pKa = 4.5TVNLPAGGSTTHH897 pKa = 6.41TASCGLSASATGTLSNTATVAAPAGVTDD925 pKa = 4.31PTPGNNSATDD935 pKa = 3.41TDD937 pKa = 4.55TITLLGAAVTASKK950 pKa = 9.37TVAGSFVPGSPVTYY964 pKa = 9.69TVTLTNSLGAQANNPGDD981 pKa = 3.91EE982 pKa = 4.4FTDD985 pKa = 3.6VLPAALTLVSATATSGSAVATIGTNTVTWNGAIPGGGSVTITITATLSPSATGTVANQGSFAYY1048 pKa = 10.4DD1049 pKa = 3.17GDD1051 pKa = 4.5ANGSNEE1057 pKa = 4.4TNGLTDD1063 pKa = 5.48DD1064 pKa = 4.44PAQPGAADD1072 pKa = 3.43PTVFTVGAAVADD1084 pKa = 4.02LAITKK1089 pKa = 9.57TDD1091 pKa = 3.23GVTTATPGGSTTYY1104 pKa = 10.45TIVASNAGPSAVTGATVTDD1123 pKa = 4.14TFPASLTCTWTCAGSGGGTCAPSGSGNIASPVTLPVGASATYY1165 pKa = 8.69TATCAISATASGTLVNTATIAVPAGVNDD1193 pKa = 3.85PTPGNNSATDD1203 pKa = 3.72TDD1205 pKa = 4.25TLAATADD1212 pKa = 3.86LAITKK1217 pKa = 9.51TDD1219 pKa = 3.38GVTSATPGDD1228 pKa = 3.83STTYY1232 pKa = 10.41TIVASNAGPSAVTGATVTDD1251 pKa = 3.96TFAASLTCTWTCAGSGGGTCAPSGSGNIASPVTLPVGASATYY1293 pKa = 8.69TATCAISATASGTLVNTATIAAPAGVTDD1321 pKa = 4.31PTPGNNSATDD1331 pKa = 3.52TDD1333 pKa = 3.84ALGAEE1338 pKa = 4.41TALSISKK1345 pKa = 9.21TDD1347 pKa = 3.12GATTVAPGGTLTYY1360 pKa = 10.01TIVASNVGPSAAVGATVSDD1379 pKa = 4.59AFPAACVAPAWTCAASGGGNCPAGGSGAIAASVDD1413 pKa = 4.05LPAGAAATFTATCPVDD1429 pKa = 3.22PALADD1434 pKa = 3.42GTVITNTATVAEE1446 pKa = 4.47AAGATDD1452 pKa = 5.28PDD1454 pKa = 4.41PSDD1457 pKa = 3.98NSATDD1462 pKa = 3.35TTTAAVPVAGAAVSVRR1478 pKa = 11.84KK1479 pKa = 9.67SVSGSFLPGGAIRR1492 pKa = 11.84YY1493 pKa = 6.61TLVLVNSGGPQGDD1506 pKa = 3.65NPGDD1510 pKa = 3.81EE1511 pKa = 4.79LTDD1514 pKa = 3.46VLPPQLVLVAASASSGAASANPATNTVTWNGAIPAGGSVTVLIDD1558 pKa = 3.07ATIRR1562 pKa = 11.84SDD1564 pKa = 3.46ASGTIGNQAGVAYY1577 pKa = 10.33DD1578 pKa = 3.73GDD1580 pKa = 4.13GDD1582 pKa = 4.14GSNEE1586 pKa = 3.7ATALSDD1592 pKa = 3.87DD1593 pKa = 4.09PAVPGAADD1601 pKa = 3.37PTVFPVGAPAVVVPVPTLTGLGALLLGLGLAGLAIAARR1639 pKa = 11.84PRR1641 pKa = 11.84RR1642 pKa = 11.84RR1643 pKa = 11.84LRR1645 pKa = 3.48
MM1 pKa = 7.39TMQSRR6 pKa = 11.84AARR9 pKa = 11.84AAVSLCVMVVALAAGALGAQPATGPLQFSAPVSAPFVEE47 pKa = 4.87TKK49 pKa = 10.4PVVANPFSEE58 pKa = 4.34AARR61 pKa = 11.84PVSPRR66 pKa = 11.84AFSTEE71 pKa = 3.73FPAEE75 pKa = 4.03VEE77 pKa = 4.39PNGTPATATPLGAATYY93 pKa = 11.27VRR95 pKa = 11.84MQGNLFPAADD105 pKa = 3.25VDD107 pKa = 4.31YY108 pKa = 11.09FSFAAQAGDD117 pKa = 4.06RR118 pKa = 11.84IYY120 pKa = 11.3AAVITAANTSTSADD134 pKa = 3.53SQLNLLDD141 pKa = 3.82TDD143 pKa = 3.78GTTVLEE149 pKa = 4.51FDD151 pKa = 5.18DD152 pKa = 6.03DD153 pKa = 4.66DD154 pKa = 5.92GSFAATSSSIAGTVIPADD172 pKa = 3.28GTYY175 pKa = 10.21YY176 pKa = 10.97LSVNHH181 pKa = 5.78FTAAQTLRR189 pKa = 11.84PYY191 pKa = 9.28EE192 pKa = 3.78LHH194 pKa = 6.69VRR196 pKa = 11.84IQRR199 pKa = 11.84GSPTPEE205 pKa = 3.74VEE207 pKa = 4.98SNDD210 pKa = 3.75TPGTANPLPANGWVSGTRR228 pKa = 11.84NPPAATEE235 pKa = 3.98QDD237 pKa = 3.29WYY239 pKa = 11.18AFTANAGDD247 pKa = 4.02TVFLSLDD254 pKa = 3.77ADD256 pKa = 3.89PEE258 pKa = 4.32RR259 pKa = 11.84DD260 pKa = 3.4GVQWDD265 pKa = 3.61GRR267 pKa = 11.84LGIALFGDD275 pKa = 4.35AANQILVVNDD285 pKa = 3.69TSSGAGNPLSEE296 pKa = 4.46ALVFTIKK303 pKa = 9.79TAGTYY308 pKa = 9.75FAFVDD313 pKa = 3.85SATAAVGGPTATYY326 pKa = 8.87HH327 pKa = 6.62LSVSVQPPPTPMPLAACTTYY347 pKa = 10.85TSTDD351 pKa = 3.19VPKK354 pKa = 10.5TIGPANGLVSSLITVPGNPRR374 pKa = 11.84IGDD377 pKa = 3.93LNVEE381 pKa = 3.99IQLNHH386 pKa = 6.73ALMADD391 pKa = 3.52VDD393 pKa = 3.68AHH395 pKa = 6.32LRR397 pKa = 11.84SPSGNDD403 pKa = 2.46IGLFTDD409 pKa = 3.65IGAAATGGQAQMDD422 pKa = 4.0VVFDD426 pKa = 4.49DD427 pKa = 4.58EE428 pKa = 4.87AAIPPTFTALRR439 pKa = 11.84NMALKK444 pKa = 10.66PEE446 pKa = 4.26LNYY449 pKa = 10.57RR450 pKa = 11.84LSWFDD455 pKa = 3.41GEE457 pKa = 4.52NAGGTWTLDD466 pKa = 3.94LRR468 pKa = 11.84DD469 pKa = 4.39DD470 pKa = 4.06VNNASGGTLTSWSLRR485 pKa = 11.84ICEE488 pKa = 4.53PPPPPTCGPGFVQTTVYY505 pKa = 9.77STDD508 pKa = 3.92FEE510 pKa = 4.57SGAAGFTHH518 pKa = 7.23SGTQDD523 pKa = 2.74EE524 pKa = 4.98WEE526 pKa = 4.66LGLPATVATTTANPVAAFNSCASGVNCWKK555 pKa = 9.61TDD557 pKa = 3.3LDD559 pKa = 3.48NTYY562 pKa = 10.8NISSNQNLTSPSINLGGFSPPIVVRR587 pKa = 11.84WSHH590 pKa = 5.1QYY592 pKa = 10.56QMEE595 pKa = 4.34SASFDD600 pKa = 3.92LYY602 pKa = 11.36SVDD605 pKa = 3.36LRR607 pKa = 11.84EE608 pKa = 4.21VGGANPVRR616 pKa = 11.84LFEE619 pKa = 4.35WLDD622 pKa = 3.3ATMTDD627 pKa = 3.2AVGNPTVNIPARR639 pKa = 11.84AGWSQVVTRR648 pKa = 11.84ADD650 pKa = 3.17ALAGQNVEE658 pKa = 4.9LNFNLTSDD666 pKa = 3.38NTVNFGGVAVDD677 pKa = 4.07DD678 pKa = 4.53VSVTACRR685 pKa = 11.84VAIADD690 pKa = 4.0LSITKK695 pKa = 9.61TDD697 pKa = 3.45GVTTAVPGGSVTYY710 pKa = 10.02TITASNAGPDD720 pKa = 3.77ADD722 pKa = 4.15TAATVADD729 pKa = 4.52TFPAALTCTWTCVGAGGGTCTAAGSGNIADD759 pKa = 4.33TVNLPSGGSVTYY771 pKa = 8.84TASCAIGPSVTGTLSNTATVTASVADD797 pKa = 4.17PNPANNSATDD807 pKa = 3.63TDD809 pKa = 4.28TLTPQADD816 pKa = 3.73LAITKK821 pKa = 9.28TDD823 pKa = 3.06GVTSVIQGGSTTYY836 pKa = 10.62TITASNAGPSNASGATVTDD855 pKa = 4.01TFPASLTCTWTCVGAGGGTCTAAGSGNIADD885 pKa = 4.5TVNLPAGGSTTHH897 pKa = 6.41TASCGLSASATGTLSNTATVAAPAGVTDD925 pKa = 4.31PTPGNNSATDD935 pKa = 3.41TDD937 pKa = 4.55TITLLGAAVTASKK950 pKa = 9.37TVAGSFVPGSPVTYY964 pKa = 9.69TVTLTNSLGAQANNPGDD981 pKa = 3.91EE982 pKa = 4.4FTDD985 pKa = 3.6VLPAALTLVSATATSGSAVATIGTNTVTWNGAIPGGGSVTITITATLSPSATGTVANQGSFAYY1048 pKa = 10.4DD1049 pKa = 3.17GDD1051 pKa = 4.5ANGSNEE1057 pKa = 4.4TNGLTDD1063 pKa = 5.48DD1064 pKa = 4.44PAQPGAADD1072 pKa = 3.43PTVFTVGAAVADD1084 pKa = 4.02LAITKK1089 pKa = 9.57TDD1091 pKa = 3.23GVTTATPGGSTTYY1104 pKa = 10.45TIVASNAGPSAVTGATVTDD1123 pKa = 4.14TFPASLTCTWTCAGSGGGTCAPSGSGNIASPVTLPVGASATYY1165 pKa = 8.69TATCAISATASGTLVNTATIAVPAGVNDD1193 pKa = 3.85PTPGNNSATDD1203 pKa = 3.72TDD1205 pKa = 4.25TLAATADD1212 pKa = 3.86LAITKK1217 pKa = 9.51TDD1219 pKa = 3.38GVTSATPGDD1228 pKa = 3.83STTYY1232 pKa = 10.41TIVASNAGPSAVTGATVTDD1251 pKa = 3.96TFAASLTCTWTCAGSGGGTCAPSGSGNIASPVTLPVGASATYY1293 pKa = 8.69TATCAISATASGTLVNTATIAAPAGVTDD1321 pKa = 4.31PTPGNNSATDD1331 pKa = 3.52TDD1333 pKa = 3.84ALGAEE1338 pKa = 4.41TALSISKK1345 pKa = 9.21TDD1347 pKa = 3.12GATTVAPGGTLTYY1360 pKa = 10.01TIVASNVGPSAAVGATVSDD1379 pKa = 4.59AFPAACVAPAWTCAASGGGNCPAGGSGAIAASVDD1413 pKa = 4.05LPAGAAATFTATCPVDD1429 pKa = 3.22PALADD1434 pKa = 3.42GTVITNTATVAEE1446 pKa = 4.47AAGATDD1452 pKa = 5.28PDD1454 pKa = 4.41PSDD1457 pKa = 3.98NSATDD1462 pKa = 3.35TTTAAVPVAGAAVSVRR1478 pKa = 11.84KK1479 pKa = 9.67SVSGSFLPGGAIRR1492 pKa = 11.84YY1493 pKa = 6.61TLVLVNSGGPQGDD1506 pKa = 3.65NPGDD1510 pKa = 3.81EE1511 pKa = 4.79LTDD1514 pKa = 3.46VLPPQLVLVAASASSGAASANPATNTVTWNGAIPAGGSVTVLIDD1558 pKa = 3.07ATIRR1562 pKa = 11.84SDD1564 pKa = 3.46ASGTIGNQAGVAYY1577 pKa = 10.33DD1578 pKa = 3.73GDD1580 pKa = 4.13GDD1582 pKa = 4.14GSNEE1586 pKa = 3.7ATALSDD1592 pKa = 3.87DD1593 pKa = 4.09PAVPGAADD1601 pKa = 3.37PTVFPVGAPAVVVPVPTLTGLGALLLGLGLAGLAIAARR1639 pKa = 11.84PRR1641 pKa = 11.84RR1642 pKa = 11.84RR1643 pKa = 11.84LRR1645 pKa = 3.48
Molecular weight: 161.99 kDa
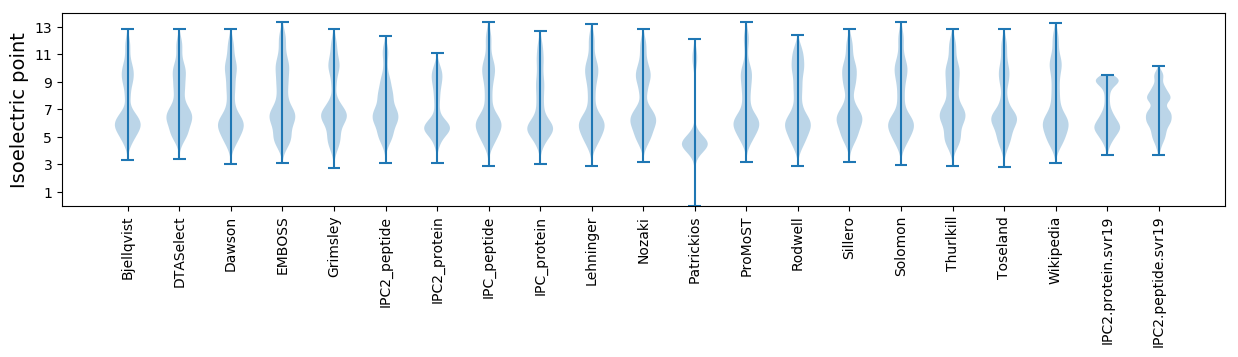
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A167H409|A0A167H409_9GAMM Uncharacterized protein OS=Dokdonella koreensis DS-123 OX=1300342 GN=I596_2808 PE=4 SV=1
MM1 pKa = 7.15GHH3 pKa = 6.7RR4 pKa = 11.84GVVFWQRR11 pKa = 11.84KK12 pKa = 4.67VRR14 pKa = 11.84GIVPGSAVRR23 pKa = 11.84AGSGRR28 pKa = 11.84FITGAQRR35 pKa = 11.84VPAAARR41 pKa = 11.84VLRR44 pKa = 11.84AA45 pKa = 3.48
MM1 pKa = 7.15GHH3 pKa = 6.7RR4 pKa = 11.84GVVFWQRR11 pKa = 11.84KK12 pKa = 4.67VRR14 pKa = 11.84GIVPGSAVRR23 pKa = 11.84AGSGRR28 pKa = 11.84FITGAQRR35 pKa = 11.84VPAAARR41 pKa = 11.84VLRR44 pKa = 11.84AA45 pKa = 3.48
Molecular weight: 4.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322314 |
37 |
2826 |
350.4 |
37.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.354 ± 0.071 | 0.855 ± 0.013 |
5.922 ± 0.028 | 4.905 ± 0.042 |
3.263 ± 0.024 | 9.031 ± 0.048 |
2.071 ± 0.021 | 4.079 ± 0.028 |
2.001 ± 0.032 | 10.811 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.707 ± 0.017 | 2.267 ± 0.028 |
5.772 ± 0.028 | 3.293 ± 0.023 |
8.218 ± 0.054 | 4.858 ± 0.043 |
5.305 ± 0.058 | 7.521 ± 0.037 |
1.496 ± 0.018 | 2.271 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
