
Jeotgalibaca porci
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae; Jeotgalibaca
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
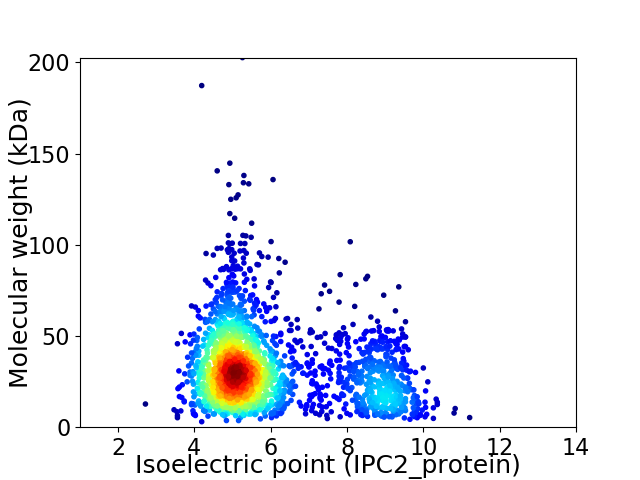
Virtual 2D-PAGE plot for 2201 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G7WF22|A0A6G7WF22_9LACT Flotillin family protein OS=Jeotgalibaca porci OX=1868793 GN=G7058_01520 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 10.46KK3 pKa = 10.23SVLSLLSTVSLLTLLTACGGGEE25 pKa = 4.14TEE27 pKa = 4.36EE28 pKa = 4.3QEE30 pKa = 4.63SVTRR34 pKa = 11.84TDD36 pKa = 3.1GTTEE40 pKa = 3.75TSEE43 pKa = 4.32TADD46 pKa = 3.44STEE49 pKa = 4.14TASGGSVYY57 pKa = 10.86YY58 pKa = 10.99LNFKK62 pKa = 10.45PEE64 pKa = 3.65IADD67 pKa = 4.31AIEE70 pKa = 4.79DD71 pKa = 3.66LAEE74 pKa = 4.7AYY76 pKa = 9.39TEE78 pKa = 4.09EE79 pKa = 4.32TGVEE83 pKa = 4.18VKK85 pKa = 9.93MVTAAGGTYY94 pKa = 9.41EE95 pKa = 4.04QTLKK99 pKa = 11.16SEE101 pKa = 4.44ITKK104 pKa = 10.16SAPPTIFFINGPIGYY119 pKa = 8.76QNWQDD124 pKa = 3.69YY125 pKa = 8.81TLDD128 pKa = 3.84LADD131 pKa = 3.59TDD133 pKa = 4.14VYY135 pKa = 11.63DD136 pKa = 3.64WLVDD140 pKa = 3.34KK141 pKa = 11.15DD142 pKa = 4.03LAITGADD149 pKa = 3.72DD150 pKa = 3.58SVAAIPITVEE160 pKa = 4.15GYY162 pKa = 10.73GIIYY166 pKa = 9.74NAAIMEE172 pKa = 4.89DD173 pKa = 4.64YY174 pKa = 10.51FALDD178 pKa = 3.24NKK180 pKa = 9.98ATDD183 pKa = 3.94FDD185 pKa = 4.62SVDD188 pKa = 4.56DD189 pKa = 4.38INNFDD194 pKa = 3.71ALKK197 pKa = 10.58EE198 pKa = 4.18VVEE201 pKa = 5.4DD202 pKa = 3.6MTALKK207 pKa = 10.37DD208 pKa = 3.54DD209 pKa = 4.81LGIQGVFSSTSLASTDD225 pKa = 2.82QWRR228 pKa = 11.84WQTHH232 pKa = 5.39LANLPVYY239 pKa = 10.31YY240 pKa = 9.8EE241 pKa = 4.0YY242 pKa = 11.08RR243 pKa = 11.84DD244 pKa = 3.64ADD246 pKa = 3.69VDD248 pKa = 3.98TMSEE252 pKa = 4.04LTFKK256 pKa = 10.91YY257 pKa = 9.81GKK259 pKa = 9.88QYY261 pKa = 11.43QNIFDD266 pKa = 5.09LYY268 pKa = 10.72INNSTNAPGLLGAKK282 pKa = 6.63TTTDD286 pKa = 2.8SMTEE290 pKa = 3.78FALGQSAMVQNGNWGWGQISGTEE313 pKa = 3.92GNVVNEE319 pKa = 4.22EE320 pKa = 4.47DD321 pKa = 3.84VHH323 pKa = 7.04FMPIYY328 pKa = 9.48TGVEE332 pKa = 3.93GEE334 pKa = 4.34EE335 pKa = 4.03NQGLAIGTEE344 pKa = 3.63NFMAVNSQASEE355 pKa = 3.93EE356 pKa = 4.71DD357 pKa = 3.22IQASLDD363 pKa = 3.91FIEE366 pKa = 4.48WMYY369 pKa = 11.44SSEE372 pKa = 4.1TGKK375 pKa = 10.63DD376 pKa = 3.36YY377 pKa = 11.42VVNQFGFIPTFNTFGEE393 pKa = 4.73DD394 pKa = 3.61EE395 pKa = 4.84FPTDD399 pKa = 4.11PLAQDD404 pKa = 3.9VLGSMNDD411 pKa = 3.13EE412 pKa = 4.51SKK414 pKa = 11.47YY415 pKa = 9.48SVPWTFTTFPSEE427 pKa = 3.83AFKK430 pKa = 11.41NDD432 pKa = 3.33FGNGLLMYY440 pKa = 10.4AQDD443 pKa = 3.26QATWDD448 pKa = 3.73EE449 pKa = 4.52VEE451 pKa = 4.42TTVVDD456 pKa = 3.99AWARR460 pKa = 11.84EE461 pKa = 3.98AALTQQQ467 pKa = 3.58
MM1 pKa = 7.35KK2 pKa = 10.46KK3 pKa = 10.23SVLSLLSTVSLLTLLTACGGGEE25 pKa = 4.14TEE27 pKa = 4.36EE28 pKa = 4.3QEE30 pKa = 4.63SVTRR34 pKa = 11.84TDD36 pKa = 3.1GTTEE40 pKa = 3.75TSEE43 pKa = 4.32TADD46 pKa = 3.44STEE49 pKa = 4.14TASGGSVYY57 pKa = 10.86YY58 pKa = 10.99LNFKK62 pKa = 10.45PEE64 pKa = 3.65IADD67 pKa = 4.31AIEE70 pKa = 4.79DD71 pKa = 3.66LAEE74 pKa = 4.7AYY76 pKa = 9.39TEE78 pKa = 4.09EE79 pKa = 4.32TGVEE83 pKa = 4.18VKK85 pKa = 9.93MVTAAGGTYY94 pKa = 9.41EE95 pKa = 4.04QTLKK99 pKa = 11.16SEE101 pKa = 4.44ITKK104 pKa = 10.16SAPPTIFFINGPIGYY119 pKa = 8.76QNWQDD124 pKa = 3.69YY125 pKa = 8.81TLDD128 pKa = 3.84LADD131 pKa = 3.59TDD133 pKa = 4.14VYY135 pKa = 11.63DD136 pKa = 3.64WLVDD140 pKa = 3.34KK141 pKa = 11.15DD142 pKa = 4.03LAITGADD149 pKa = 3.72DD150 pKa = 3.58SVAAIPITVEE160 pKa = 4.15GYY162 pKa = 10.73GIIYY166 pKa = 9.74NAAIMEE172 pKa = 4.89DD173 pKa = 4.64YY174 pKa = 10.51FALDD178 pKa = 3.24NKK180 pKa = 9.98ATDD183 pKa = 3.94FDD185 pKa = 4.62SVDD188 pKa = 4.56DD189 pKa = 4.38INNFDD194 pKa = 3.71ALKK197 pKa = 10.58EE198 pKa = 4.18VVEE201 pKa = 5.4DD202 pKa = 3.6MTALKK207 pKa = 10.37DD208 pKa = 3.54DD209 pKa = 4.81LGIQGVFSSTSLASTDD225 pKa = 2.82QWRR228 pKa = 11.84WQTHH232 pKa = 5.39LANLPVYY239 pKa = 10.31YY240 pKa = 9.8EE241 pKa = 4.0YY242 pKa = 11.08RR243 pKa = 11.84DD244 pKa = 3.64ADD246 pKa = 3.69VDD248 pKa = 3.98TMSEE252 pKa = 4.04LTFKK256 pKa = 10.91YY257 pKa = 9.81GKK259 pKa = 9.88QYY261 pKa = 11.43QNIFDD266 pKa = 5.09LYY268 pKa = 10.72INNSTNAPGLLGAKK282 pKa = 6.63TTTDD286 pKa = 2.8SMTEE290 pKa = 3.78FALGQSAMVQNGNWGWGQISGTEE313 pKa = 3.92GNVVNEE319 pKa = 4.22EE320 pKa = 4.47DD321 pKa = 3.84VHH323 pKa = 7.04FMPIYY328 pKa = 9.48TGVEE332 pKa = 3.93GEE334 pKa = 4.34EE335 pKa = 4.03NQGLAIGTEE344 pKa = 3.63NFMAVNSQASEE355 pKa = 3.93EE356 pKa = 4.71DD357 pKa = 3.22IQASLDD363 pKa = 3.91FIEE366 pKa = 4.48WMYY369 pKa = 11.44SSEE372 pKa = 4.1TGKK375 pKa = 10.63DD376 pKa = 3.36YY377 pKa = 11.42VVNQFGFIPTFNTFGEE393 pKa = 4.73DD394 pKa = 3.61EE395 pKa = 4.84FPTDD399 pKa = 4.11PLAQDD404 pKa = 3.9VLGSMNDD411 pKa = 3.13EE412 pKa = 4.51SKK414 pKa = 11.47YY415 pKa = 9.48SVPWTFTTFPSEE427 pKa = 3.83AFKK430 pKa = 11.41NDD432 pKa = 3.33FGNGLLMYY440 pKa = 10.4AQDD443 pKa = 3.26QATWDD448 pKa = 3.73EE449 pKa = 4.52VEE451 pKa = 4.42TTVVDD456 pKa = 3.99AWARR460 pKa = 11.84EE461 pKa = 3.98AALTQQQ467 pKa = 3.58
Molecular weight: 51.44 kDa
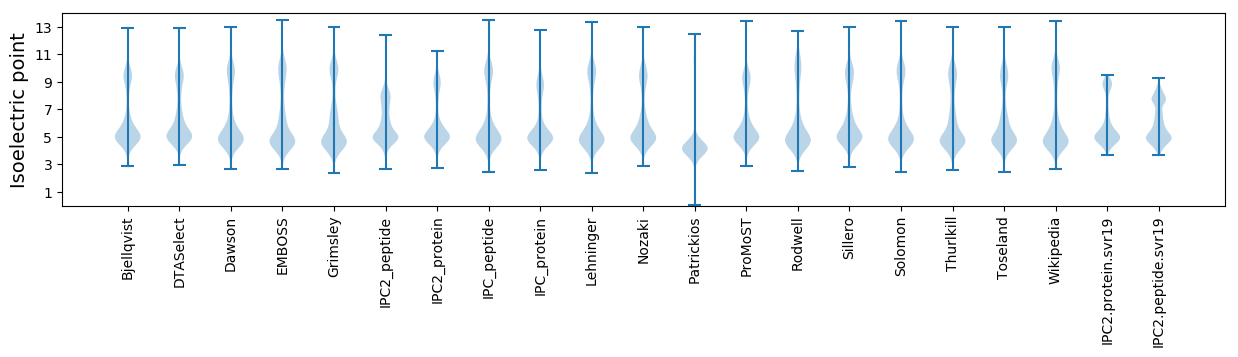
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G7WH95|A0A6G7WH95_9LACT Flagellar hook-associated protein 2 OS=Jeotgalibaca porci OX=1868793 GN=fliD PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.32KK9 pKa = 9.49RR10 pKa = 11.84KK11 pKa = 8.83RR12 pKa = 11.84QTVHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MSTKK25 pKa = 10.02NGRR28 pKa = 11.84NVLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.73GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.32KK9 pKa = 9.49RR10 pKa = 11.84KK11 pKa = 8.83RR12 pKa = 11.84QTVHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MSTKK25 pKa = 10.02NGRR28 pKa = 11.84NVLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.73GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.05
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
663762 |
27 |
1756 |
301.6 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.427 ± 0.05 | 0.531 ± 0.013 |
5.459 ± 0.039 | 7.759 ± 0.062 |
4.446 ± 0.041 | 6.658 ± 0.046 |
1.927 ± 0.025 | 7.62 ± 0.052 |
6.252 ± 0.045 | 9.59 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.891 ± 0.025 | 4.642 ± 0.035 |
3.476 ± 0.025 | 3.68 ± 0.034 |
4.186 ± 0.036 | 5.794 ± 0.033 |
5.981 ± 0.038 | 7.042 ± 0.042 |
0.921 ± 0.02 | 3.717 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
