
Escovopsis weberi
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Hypocreaceae; Escovopsis
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
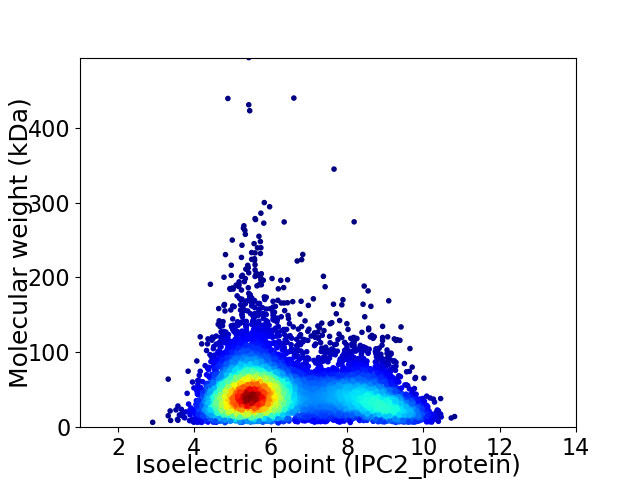
Virtual 2D-PAGE plot for 6869 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M8N3D9|A0A0M8N3D9_9HYPO Uncharacterized protein OS=Escovopsis weberi OX=150374 GN=ESCO_001134 PE=4 SV=1
MM1 pKa = 7.79DD2 pKa = 3.55VCTTSQQNGFNWSGLAPGEE21 pKa = 4.19VGSYY25 pKa = 11.01DD26 pKa = 3.38GFKK29 pKa = 10.77FKK31 pKa = 10.21GWKK34 pKa = 10.42CEE36 pKa = 4.14DD37 pKa = 3.34NQADD41 pKa = 3.85VNINALGLNVNVGLGLGGSGAQTARR66 pKa = 11.84TIVGDD71 pKa = 3.76LTSNTDD77 pKa = 3.51DD78 pKa = 4.25SPSISADD85 pKa = 3.2GPFSIDD91 pKa = 2.96EE92 pKa = 4.35MQVSTEE98 pKa = 3.71FDD100 pKa = 3.13ADD102 pKa = 3.7LEE104 pKa = 4.39FRR106 pKa = 11.84YY107 pKa = 10.67EE108 pKa = 4.05MADD111 pKa = 2.9GSYY114 pKa = 10.34CMHH117 pKa = 6.58VASCSPSGTTVKK129 pKa = 9.5NTGCHH134 pKa = 5.35GSKK137 pKa = 10.53GVTVVYY143 pKa = 9.96PGSSGSNDD151 pKa = 2.95SCRR154 pKa = 11.84TRR156 pKa = 11.84IHH158 pKa = 7.36GITFNCPNGSSPSGGQQPSQEE179 pKa = 4.22PAQEE183 pKa = 4.15PSQAPTQAPTEE194 pKa = 4.08QPTQVSHH201 pKa = 6.62TSAPDD206 pKa = 3.29ASSTDD211 pKa = 3.6SPVSSGGDD219 pKa = 3.31DD220 pKa = 6.19DD221 pKa = 5.21EE222 pKa = 7.24DD223 pKa = 5.31CDD225 pKa = 3.97EE226 pKa = 4.51TTTAPASAPTALHH239 pKa = 6.67PLRR242 pKa = 11.84PFIPHH247 pKa = 7.14PGADD251 pKa = 3.56ASSDD255 pKa = 3.46APAASSAPATSLRR268 pKa = 11.84IPLHH272 pKa = 5.54TGVPVRR278 pKa = 11.84VPTALPGDD286 pKa = 4.22STTAPAEE293 pKa = 4.17PTSVAPAEE301 pKa = 4.28PANTEE306 pKa = 3.93APAEE310 pKa = 4.18PTSEE314 pKa = 4.35APAEE318 pKa = 4.04PTPTEE323 pKa = 4.19DD324 pKa = 3.89VPAQEE329 pKa = 4.82PVSTDD334 pKa = 3.0APALPEE340 pKa = 4.19STDD343 pKa = 3.42APLEE347 pKa = 4.32TVTTTFQSISTIFTTSIHH365 pKa = 6.72TITSCAPDD373 pKa = 3.61VTDD376 pKa = 4.54CPARR380 pKa = 11.84TSPTVVTEE388 pKa = 4.25TVVHH392 pKa = 6.7DD393 pKa = 3.71TTICPVTATLTRR405 pKa = 11.84TRR407 pKa = 11.84AHH409 pKa = 6.55HH410 pKa = 5.94SAPSAHH416 pKa = 5.93TRR418 pKa = 11.84VAPPTDD424 pKa = 3.7AAPTSIPTQDD434 pKa = 3.03TAAPEE439 pKa = 4.35APAPTSVAPGTGAPEE454 pKa = 4.15APAPTSAAPGTDD466 pKa = 3.7APDD469 pKa = 3.69APAPTSAAPGTDD481 pKa = 3.7APDD484 pKa = 3.58APAPTGAAPGTDD496 pKa = 3.88APDD499 pKa = 4.15APDD502 pKa = 3.86APAPTGAAPGTDD514 pKa = 3.56APEE517 pKa = 4.44APAPTSAAPGTDD529 pKa = 3.26APSSPEE535 pKa = 3.98SASPSAPAEE544 pKa = 4.28AEE546 pKa = 4.14PTSAPAEE553 pKa = 4.41TFVTTYY559 pKa = 10.84QSTSTAYY566 pKa = 8.08VTQVHH571 pKa = 7.02TITSCAPDD579 pKa = 4.04VPDD582 pKa = 4.25CPARR586 pKa = 11.84AGPTVVTEE594 pKa = 4.49TVAVSTTICPVTATMTRR611 pKa = 11.84PAATPSAPAAPVGAPAPTGAPQGSDD636 pKa = 3.15APASPVDD643 pKa = 3.93VPAPADD649 pKa = 3.71SPAPAPAPEE658 pKa = 4.93GSPAPAPDD666 pKa = 4.03APAPAPDD673 pKa = 3.8APAPEE678 pKa = 5.34DD679 pKa = 3.66SPAPADD685 pKa = 3.84APASPEE691 pKa = 3.68NSAPAVIYY699 pKa = 10.74SPVPTTMQSAPGPAGPTAFVPQPTNDD725 pKa = 3.44LPGPPVPTGMPVISGAGRR743 pKa = 11.84LGASAGLLLGLMAAVVLLL761 pKa = 4.07
MM1 pKa = 7.79DD2 pKa = 3.55VCTTSQQNGFNWSGLAPGEE21 pKa = 4.19VGSYY25 pKa = 11.01DD26 pKa = 3.38GFKK29 pKa = 10.77FKK31 pKa = 10.21GWKK34 pKa = 10.42CEE36 pKa = 4.14DD37 pKa = 3.34NQADD41 pKa = 3.85VNINALGLNVNVGLGLGGSGAQTARR66 pKa = 11.84TIVGDD71 pKa = 3.76LTSNTDD77 pKa = 3.51DD78 pKa = 4.25SPSISADD85 pKa = 3.2GPFSIDD91 pKa = 2.96EE92 pKa = 4.35MQVSTEE98 pKa = 3.71FDD100 pKa = 3.13ADD102 pKa = 3.7LEE104 pKa = 4.39FRR106 pKa = 11.84YY107 pKa = 10.67EE108 pKa = 4.05MADD111 pKa = 2.9GSYY114 pKa = 10.34CMHH117 pKa = 6.58VASCSPSGTTVKK129 pKa = 9.5NTGCHH134 pKa = 5.35GSKK137 pKa = 10.53GVTVVYY143 pKa = 9.96PGSSGSNDD151 pKa = 2.95SCRR154 pKa = 11.84TRR156 pKa = 11.84IHH158 pKa = 7.36GITFNCPNGSSPSGGQQPSQEE179 pKa = 4.22PAQEE183 pKa = 4.15PSQAPTQAPTEE194 pKa = 4.08QPTQVSHH201 pKa = 6.62TSAPDD206 pKa = 3.29ASSTDD211 pKa = 3.6SPVSSGGDD219 pKa = 3.31DD220 pKa = 6.19DD221 pKa = 5.21EE222 pKa = 7.24DD223 pKa = 5.31CDD225 pKa = 3.97EE226 pKa = 4.51TTTAPASAPTALHH239 pKa = 6.67PLRR242 pKa = 11.84PFIPHH247 pKa = 7.14PGADD251 pKa = 3.56ASSDD255 pKa = 3.46APAASSAPATSLRR268 pKa = 11.84IPLHH272 pKa = 5.54TGVPVRR278 pKa = 11.84VPTALPGDD286 pKa = 4.22STTAPAEE293 pKa = 4.17PTSVAPAEE301 pKa = 4.28PANTEE306 pKa = 3.93APAEE310 pKa = 4.18PTSEE314 pKa = 4.35APAEE318 pKa = 4.04PTPTEE323 pKa = 4.19DD324 pKa = 3.89VPAQEE329 pKa = 4.82PVSTDD334 pKa = 3.0APALPEE340 pKa = 4.19STDD343 pKa = 3.42APLEE347 pKa = 4.32TVTTTFQSISTIFTTSIHH365 pKa = 6.72TITSCAPDD373 pKa = 3.61VTDD376 pKa = 4.54CPARR380 pKa = 11.84TSPTVVTEE388 pKa = 4.25TVVHH392 pKa = 6.7DD393 pKa = 3.71TTICPVTATLTRR405 pKa = 11.84TRR407 pKa = 11.84AHH409 pKa = 6.55HH410 pKa = 5.94SAPSAHH416 pKa = 5.93TRR418 pKa = 11.84VAPPTDD424 pKa = 3.7AAPTSIPTQDD434 pKa = 3.03TAAPEE439 pKa = 4.35APAPTSVAPGTGAPEE454 pKa = 4.15APAPTSAAPGTDD466 pKa = 3.7APDD469 pKa = 3.69APAPTSAAPGTDD481 pKa = 3.7APDD484 pKa = 3.58APAPTGAAPGTDD496 pKa = 3.88APDD499 pKa = 4.15APDD502 pKa = 3.86APAPTGAAPGTDD514 pKa = 3.56APEE517 pKa = 4.44APAPTSAAPGTDD529 pKa = 3.26APSSPEE535 pKa = 3.98SASPSAPAEE544 pKa = 4.28AEE546 pKa = 4.14PTSAPAEE553 pKa = 4.41TFVTTYY559 pKa = 10.84QSTSTAYY566 pKa = 8.08VTQVHH571 pKa = 7.02TITSCAPDD579 pKa = 4.04VPDD582 pKa = 4.25CPARR586 pKa = 11.84AGPTVVTEE594 pKa = 4.49TVAVSTTICPVTATMTRR611 pKa = 11.84PAATPSAPAAPVGAPAPTGAPQGSDD636 pKa = 3.15APASPVDD643 pKa = 3.93VPAPADD649 pKa = 3.71SPAPAPAPEE658 pKa = 4.93GSPAPAPDD666 pKa = 4.03APAPAPDD673 pKa = 3.8APAPEE678 pKa = 5.34DD679 pKa = 3.66SPAPADD685 pKa = 3.84APASPEE691 pKa = 3.68NSAPAVIYY699 pKa = 10.74SPVPTTMQSAPGPAGPTAFVPQPTNDD725 pKa = 3.44LPGPPVPTGMPVISGAGRR743 pKa = 11.84LGASAGLLLGLMAAVVLLL761 pKa = 4.07
Molecular weight: 74.8 kDa
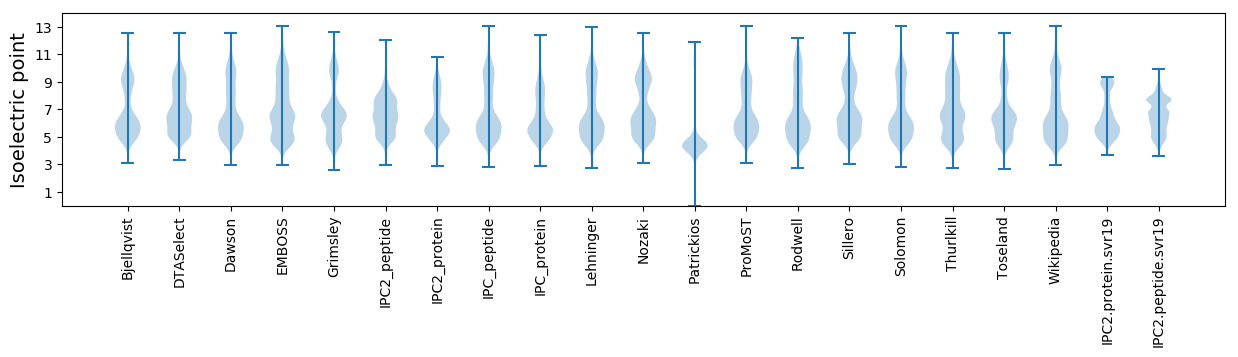
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M8MZG8|A0A0M8MZG8_9HYPO Uncharacterized protein OS=Escovopsis weberi OX=150374 GN=ESCO_005626 PE=4 SV=1
MM1 pKa = 7.52SFLARR6 pKa = 11.84LARR9 pKa = 11.84PVVALHH15 pKa = 6.26IQPSTRR21 pKa = 11.84TFTTLQPLRR30 pKa = 11.84PTLLATFRR38 pKa = 11.84PALTSFAPATPSTAAPGAGEE58 pKa = 4.02RR59 pKa = 11.84AADD62 pKa = 4.31LVPAAAVSAHH72 pKa = 6.75PALAGATQIRR82 pKa = 11.84FGPRR86 pKa = 11.84NTMQGHH92 pKa = 5.07TRR94 pKa = 11.84LVQKK98 pKa = 10.38RR99 pKa = 11.84RR100 pKa = 11.84HH101 pKa = 4.91GWLKK105 pKa = 10.59RR106 pKa = 11.84MRR108 pKa = 11.84SKK110 pKa = 9.65TGRR113 pKa = 11.84RR114 pKa = 11.84ILEE117 pKa = 3.75RR118 pKa = 11.84RR119 pKa = 11.84KK120 pKa = 10.16LKK122 pKa = 10.35GRR124 pKa = 11.84RR125 pKa = 11.84HH126 pKa = 4.95VAWW129 pKa = 4.03
MM1 pKa = 7.52SFLARR6 pKa = 11.84LARR9 pKa = 11.84PVVALHH15 pKa = 6.26IQPSTRR21 pKa = 11.84TFTTLQPLRR30 pKa = 11.84PTLLATFRR38 pKa = 11.84PALTSFAPATPSTAAPGAGEE58 pKa = 4.02RR59 pKa = 11.84AADD62 pKa = 4.31LVPAAAVSAHH72 pKa = 6.75PALAGATQIRR82 pKa = 11.84FGPRR86 pKa = 11.84NTMQGHH92 pKa = 5.07TRR94 pKa = 11.84LVQKK98 pKa = 10.38RR99 pKa = 11.84RR100 pKa = 11.84HH101 pKa = 4.91GWLKK105 pKa = 10.59RR106 pKa = 11.84MRR108 pKa = 11.84SKK110 pKa = 9.65TGRR113 pKa = 11.84RR114 pKa = 11.84ILEE117 pKa = 3.75RR118 pKa = 11.84RR119 pKa = 11.84KK120 pKa = 10.16LKK122 pKa = 10.35GRR124 pKa = 11.84RR125 pKa = 11.84HH126 pKa = 4.95VAWW129 pKa = 4.03
Molecular weight: 14.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3220148 |
50 |
4362 |
468.8 |
51.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.003 ± 0.032 | 1.152 ± 0.011 |
6.128 ± 0.021 | 6.321 ± 0.031 |
3.591 ± 0.018 | 7.325 ± 0.031 |
2.435 ± 0.012 | 4.434 ± 0.019 |
4.614 ± 0.026 | 8.891 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.248 ± 0.011 | 3.258 ± 0.016 |
5.96 ± 0.032 | 3.965 ± 0.026 |
6.675 ± 0.027 | 7.747 ± 0.028 |
5.301 ± 0.019 | 6.11 ± 0.024 |
1.37 ± 0.011 | 2.464 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
