
Macaca mulatta feces associated virus 9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; unclassified Porprismacovirus
Average proteome isoelectric point is 8.2
Get precalculated fractions of proteins
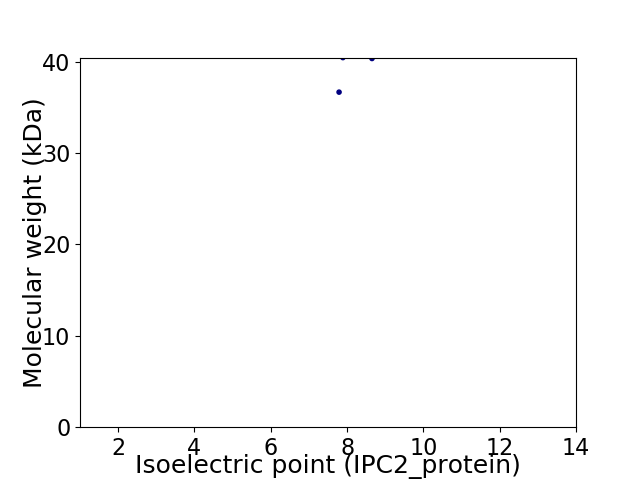
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5PZN0|A0A1W5PZN0_9VIRU Cap OS=Macaca mulatta feces associated virus 9 OX=2499231 PE=4 SV=1
MM1 pKa = 6.77MSYY4 pKa = 10.33RR5 pKa = 11.84KK6 pKa = 9.54RR7 pKa = 11.84RR8 pKa = 11.84GSKK11 pKa = 9.55VLHH14 pKa = 6.49QKK16 pKa = 8.52FQWFFDD22 pKa = 3.94LQTKK26 pKa = 9.57ADD28 pKa = 3.7EE29 pKa = 4.12MQIIEE34 pKa = 4.4VSAGGYY40 pKa = 9.27GVYY43 pKa = 10.2KK44 pKa = 10.55RR45 pKa = 11.84LFPFFSAFKK54 pKa = 8.75YY55 pKa = 10.37YY56 pKa = 10.95KK57 pKa = 10.56LGGVKK62 pKa = 9.37MKK64 pKa = 9.33MIPASTLPVDD74 pKa = 3.87PTGLSYY80 pKa = 10.68EE81 pKa = 4.23AGEE84 pKa = 4.26NTVDD88 pKa = 4.85PRR90 pKa = 11.84DD91 pKa = 3.48QLTPGLTRR99 pKa = 11.84ITNGEE104 pKa = 4.0DD105 pKa = 3.58VYY107 pKa = 11.46TKK109 pKa = 11.0LDD111 pKa = 3.67GLTGNQQRR119 pKa = 11.84EE120 pKa = 4.68LYY122 pKa = 9.13EE123 pKa = 4.83SMMIDD128 pKa = 3.25QRR130 pKa = 11.84WFKK133 pKa = 10.13WQLQSGLSRR142 pKa = 11.84YY143 pKa = 8.4ARR145 pKa = 11.84PMYY148 pKa = 8.75WQIGQLHH155 pKa = 6.12QDD157 pKa = 3.76YY158 pKa = 11.0LPGSVRR164 pKa = 11.84NLADD168 pKa = 3.42TNLTEE173 pKa = 4.61DD174 pKa = 4.17CQSISAVYY182 pKa = 10.49DD183 pKa = 3.36GANPSGPSAGPITVAADD200 pKa = 3.17ASDD203 pKa = 3.55PRR205 pKa = 11.84GLFQVGHH212 pKa = 7.01RR213 pKa = 11.84GRR215 pKa = 11.84LGWMPTDD222 pKa = 3.53GVLLKK227 pKa = 9.81GTSGGDD233 pKa = 3.59GTVHH237 pKa = 5.98AQAVEE242 pKa = 3.92AAIPAVNVFTIVTPPMHH259 pKa = 6.15KK260 pKa = 8.5TNYY263 pKa = 7.64YY264 pKa = 8.29YY265 pKa = 10.56RR266 pKa = 11.84VFVTEE271 pKa = 4.34DD272 pKa = 3.84VYY274 pKa = 11.28FKK276 pKa = 11.2SPVVVGYY283 pKa = 10.0QNFRR287 pKa = 11.84SIDD290 pKa = 3.53RR291 pKa = 11.84FVQPQFPVAKK301 pKa = 9.97LPTVNSATTDD311 pKa = 3.39SNKK314 pKa = 9.75PFPTNDD320 pKa = 3.19GGEE323 pKa = 4.25LPGVIQQ329 pKa = 4.56
MM1 pKa = 6.77MSYY4 pKa = 10.33RR5 pKa = 11.84KK6 pKa = 9.54RR7 pKa = 11.84RR8 pKa = 11.84GSKK11 pKa = 9.55VLHH14 pKa = 6.49QKK16 pKa = 8.52FQWFFDD22 pKa = 3.94LQTKK26 pKa = 9.57ADD28 pKa = 3.7EE29 pKa = 4.12MQIIEE34 pKa = 4.4VSAGGYY40 pKa = 9.27GVYY43 pKa = 10.2KK44 pKa = 10.55RR45 pKa = 11.84LFPFFSAFKK54 pKa = 8.75YY55 pKa = 10.37YY56 pKa = 10.95KK57 pKa = 10.56LGGVKK62 pKa = 9.37MKK64 pKa = 9.33MIPASTLPVDD74 pKa = 3.87PTGLSYY80 pKa = 10.68EE81 pKa = 4.23AGEE84 pKa = 4.26NTVDD88 pKa = 4.85PRR90 pKa = 11.84DD91 pKa = 3.48QLTPGLTRR99 pKa = 11.84ITNGEE104 pKa = 4.0DD105 pKa = 3.58VYY107 pKa = 11.46TKK109 pKa = 11.0LDD111 pKa = 3.67GLTGNQQRR119 pKa = 11.84EE120 pKa = 4.68LYY122 pKa = 9.13EE123 pKa = 4.83SMMIDD128 pKa = 3.25QRR130 pKa = 11.84WFKK133 pKa = 10.13WQLQSGLSRR142 pKa = 11.84YY143 pKa = 8.4ARR145 pKa = 11.84PMYY148 pKa = 8.75WQIGQLHH155 pKa = 6.12QDD157 pKa = 3.76YY158 pKa = 11.0LPGSVRR164 pKa = 11.84NLADD168 pKa = 3.42TNLTEE173 pKa = 4.61DD174 pKa = 4.17CQSISAVYY182 pKa = 10.49DD183 pKa = 3.36GANPSGPSAGPITVAADD200 pKa = 3.17ASDD203 pKa = 3.55PRR205 pKa = 11.84GLFQVGHH212 pKa = 7.01RR213 pKa = 11.84GRR215 pKa = 11.84LGWMPTDD222 pKa = 3.53GVLLKK227 pKa = 9.81GTSGGDD233 pKa = 3.59GTVHH237 pKa = 5.98AQAVEE242 pKa = 3.92AAIPAVNVFTIVTPPMHH259 pKa = 6.15KK260 pKa = 8.5TNYY263 pKa = 7.64YY264 pKa = 8.29YY265 pKa = 10.56RR266 pKa = 11.84VFVTEE271 pKa = 4.34DD272 pKa = 3.84VYY274 pKa = 11.28FKK276 pKa = 11.2SPVVVGYY283 pKa = 10.0QNFRR287 pKa = 11.84SIDD290 pKa = 3.53RR291 pKa = 11.84FVQPQFPVAKK301 pKa = 9.97LPTVNSATTDD311 pKa = 3.39SNKK314 pKa = 9.75PFPTNDD320 pKa = 3.19GGEE323 pKa = 4.25LPGVIQQ329 pKa = 4.56
Molecular weight: 36.67 kDa
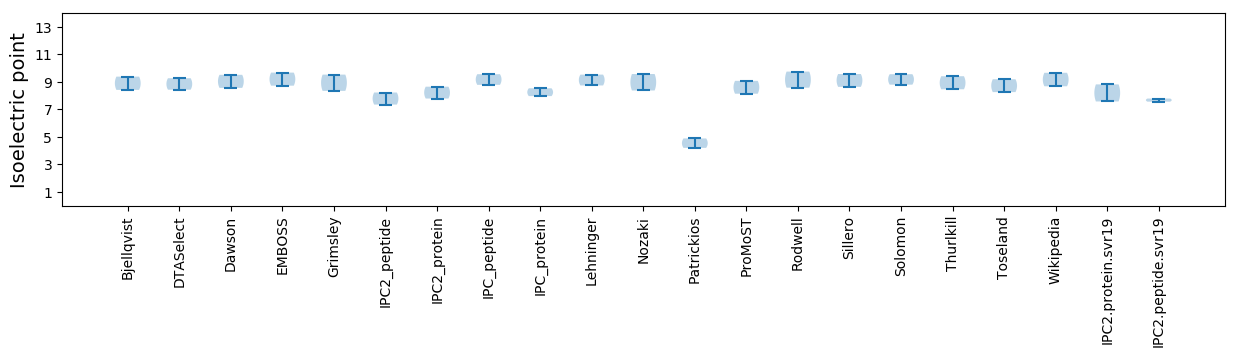
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5PZN0|A0A1W5PZN0_9VIRU Cap OS=Macaca mulatta feces associated virus 9 OX=2499231 PE=4 SV=1
MM1 pKa = 7.2CWLYY5 pKa = 11.25KK6 pKa = 10.07FAPKK10 pKa = 9.93PIPAPARR17 pKa = 11.84VDD19 pKa = 3.22KK20 pKa = 11.03ASRR23 pKa = 11.84GSINSASRR31 pKa = 11.84NSYY34 pKa = 9.71HH35 pKa = 6.89ICHH38 pKa = 6.32TCEE41 pKa = 3.69TFRR44 pKa = 11.84GCGSLSCSTLFRR56 pKa = 11.84TSLFITVQHH65 pKa = 5.32CTIMYY70 pKa = 7.67MAAQTNYY77 pKa = 10.39KK78 pKa = 9.6FRR80 pKa = 11.84QRR82 pKa = 11.84LQSIMTHH89 pKa = 4.25TWMLTVPRR97 pKa = 11.84RR98 pKa = 11.84KK99 pKa = 7.6TARR102 pKa = 11.84EE103 pKa = 3.42WMGISKK109 pKa = 9.41WLRR112 pKa = 11.84DD113 pKa = 3.56NDD115 pKa = 3.66VHH117 pKa = 6.67KK118 pKa = 9.25WTVAMEE124 pKa = 4.39TGDD127 pKa = 3.25NGYY130 pKa = 9.67DD131 pKa = 2.78HH132 pKa = 6.76WQIRR136 pKa = 11.84LQVNKK141 pKa = 9.04TFKK144 pKa = 10.1QLKK147 pKa = 9.6KK148 pKa = 8.91EE149 pKa = 4.17WGPKK153 pKa = 9.16AHH155 pKa = 6.94IEE157 pKa = 4.11EE158 pKa = 5.74ASDD161 pKa = 2.87TWNYY165 pKa = 8.1EE166 pKa = 3.95RR167 pKa = 11.84KK168 pKa = 9.95SGLFFSSNDD177 pKa = 3.29TPEE180 pKa = 3.66VRR182 pKa = 11.84KK183 pKa = 10.19CRR185 pKa = 11.84FGRR188 pKa = 11.84LTWRR192 pKa = 11.84QQAVVQAVQGSNDD205 pKa = 3.33RR206 pKa = 11.84QIVVWYY212 pKa = 9.66DD213 pKa = 3.15PNGNAGKK220 pKa = 9.94SWLLGHH226 pKa = 7.53LYY228 pKa = 8.31EE229 pKa = 5.2TGQAWVVQAQDD240 pKa = 3.45TVKK243 pKa = 11.18GIIQDD248 pKa = 3.78CASEE252 pKa = 4.39YY253 pKa = 9.83IQHH256 pKa = 5.81GWRR259 pKa = 11.84PFVVIDD265 pKa = 5.11IPRR268 pKa = 11.84SWKK271 pKa = 7.84WTQEE275 pKa = 3.77LYY277 pKa = 10.93VALEE281 pKa = 4.21RR282 pKa = 11.84IKK284 pKa = 11.05DD285 pKa = 3.81GLIKK289 pKa = 10.46DD290 pKa = 3.65PRR292 pKa = 11.84YY293 pKa = 10.2GSKK296 pKa = 8.91TVHH299 pKa = 5.15IRR301 pKa = 11.84GVKK304 pKa = 10.11VLVTCNTKK312 pKa = 10.43PKK314 pKa = 10.02LDD316 pKa = 3.92KK317 pKa = 10.98LSADD321 pKa = 3.09RR322 pKa = 11.84WVIMNMNTLEE332 pKa = 4.27AEE334 pKa = 4.33CAAMEE339 pKa = 4.18VADD342 pKa = 6.46RR343 pKa = 11.84IWEE346 pKa = 4.1DD347 pKa = 2.99
MM1 pKa = 7.2CWLYY5 pKa = 11.25KK6 pKa = 10.07FAPKK10 pKa = 9.93PIPAPARR17 pKa = 11.84VDD19 pKa = 3.22KK20 pKa = 11.03ASRR23 pKa = 11.84GSINSASRR31 pKa = 11.84NSYY34 pKa = 9.71HH35 pKa = 6.89ICHH38 pKa = 6.32TCEE41 pKa = 3.69TFRR44 pKa = 11.84GCGSLSCSTLFRR56 pKa = 11.84TSLFITVQHH65 pKa = 5.32CTIMYY70 pKa = 7.67MAAQTNYY77 pKa = 10.39KK78 pKa = 9.6FRR80 pKa = 11.84QRR82 pKa = 11.84LQSIMTHH89 pKa = 4.25TWMLTVPRR97 pKa = 11.84RR98 pKa = 11.84KK99 pKa = 7.6TARR102 pKa = 11.84EE103 pKa = 3.42WMGISKK109 pKa = 9.41WLRR112 pKa = 11.84DD113 pKa = 3.56NDD115 pKa = 3.66VHH117 pKa = 6.67KK118 pKa = 9.25WTVAMEE124 pKa = 4.39TGDD127 pKa = 3.25NGYY130 pKa = 9.67DD131 pKa = 2.78HH132 pKa = 6.76WQIRR136 pKa = 11.84LQVNKK141 pKa = 9.04TFKK144 pKa = 10.1QLKK147 pKa = 9.6KK148 pKa = 8.91EE149 pKa = 4.17WGPKK153 pKa = 9.16AHH155 pKa = 6.94IEE157 pKa = 4.11EE158 pKa = 5.74ASDD161 pKa = 2.87TWNYY165 pKa = 8.1EE166 pKa = 3.95RR167 pKa = 11.84KK168 pKa = 9.95SGLFFSSNDD177 pKa = 3.29TPEE180 pKa = 3.66VRR182 pKa = 11.84KK183 pKa = 10.19CRR185 pKa = 11.84FGRR188 pKa = 11.84LTWRR192 pKa = 11.84QQAVVQAVQGSNDD205 pKa = 3.33RR206 pKa = 11.84QIVVWYY212 pKa = 9.66DD213 pKa = 3.15PNGNAGKK220 pKa = 9.94SWLLGHH226 pKa = 7.53LYY228 pKa = 8.31EE229 pKa = 5.2TGQAWVVQAQDD240 pKa = 3.45TVKK243 pKa = 11.18GIIQDD248 pKa = 3.78CASEE252 pKa = 4.39YY253 pKa = 9.83IQHH256 pKa = 5.81GWRR259 pKa = 11.84PFVVIDD265 pKa = 5.11IPRR268 pKa = 11.84SWKK271 pKa = 7.84WTQEE275 pKa = 3.77LYY277 pKa = 10.93VALEE281 pKa = 4.21RR282 pKa = 11.84IKK284 pKa = 11.05DD285 pKa = 3.81GLIKK289 pKa = 10.46DD290 pKa = 3.65PRR292 pKa = 11.84YY293 pKa = 10.2GSKK296 pKa = 8.91TVHH299 pKa = 5.15IRR301 pKa = 11.84GVKK304 pKa = 10.11VLVTCNTKK312 pKa = 10.43PKK314 pKa = 10.02LDD316 pKa = 3.92KK317 pKa = 10.98LSADD321 pKa = 3.09RR322 pKa = 11.84WVIMNMNTLEE332 pKa = 4.27AEE334 pKa = 4.33CAAMEE339 pKa = 4.18VADD342 pKa = 6.46RR343 pKa = 11.84IWEE346 pKa = 4.1DD347 pKa = 2.99
Molecular weight: 40.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
676 |
329 |
347 |
338.0 |
38.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.361 ± 0.177 | 1.627 ± 0.832 |
5.621 ± 0.288 | 3.994 ± 0.409 |
3.846 ± 0.639 | 7.396 ± 1.273 |
2.219 ± 0.439 | 4.734 ± 0.683 |
5.917 ± 0.662 | 6.509 ± 0.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.959 ± 0.051 | 3.846 ± 0.125 |
5.178 ± 1.14 | 5.621 ± 0.288 |
6.213 ± 0.657 | 6.065 ± 0.009 |
6.953 ± 0.024 | 7.544 ± 0.416 |
3.254 ± 1.09 | 4.142 ± 0.644 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
